Plant supplies
For producing overexpression strains of TaTCP6 (TaTCP6-OE) and TaSPX1 (TaSPX1-OE), the full-length coding sequence (CDS) of TaTCP6-B and TaSPX1-D fused with 3×Flag tag sequence was cloned into the PLC41-Z2 vector respectively to generate Ubipro::TaTCP6-B-Flag and Ubipro::TaSPX1-D-Flag. To generate the fabric of TaTCP6-SRDX, the full-length coding sequence (CDS) of TaTCP6-B and its 3K promoter fused with SRDX tag sequence was cloned into the PLC41-Z2 vector to generate TaTCP6-Bpro::TaTCP6-B-SRDX. For producing overexpression strains of TaPHR2 (TaPHR2-OE), the full-length coding sequence (CDS) of TaPHR2-B fused with GFP tag sequence was cloned into the PLC41-Z2 vector respectively to generate Ubipro::TaPHR2-B-GFP. These vectors had been additional remodeled into wheat cultivar “Fielder” respectively through Agrobacterium tumefaciens-mediated gene transformation. The Tatcp6 mutant crops had been generated by CRISPR/Cas9-based gene enhancing. In short, the sgRNA goal sequences for TaTCP6-A/B/D gene enhancing had been first designed utilizing the web-based CRISPR-Cereal (http://crispr.hzau.edu.cn/CRISPR-Cereal/). Then, the sgRNA sequences had been launched into the pBUE414 vector through T4 DNA ligase (M0202, NEB). The ensuing constructs had been transferred into the A. tumefaciens pressure EHA105 and remodeled into wheat cultivar “Fielder” by A. tumefaciens-mediated gene transformation. All these wheat supplies had been validated by PCR and sequencing. The primers used on this research are listed in Supplementary Knowledge 12.
Development circumstances and strategies of discipline experiment
The sphere experiments for wheat rising had been performed within the Shangzhuang Experimental Station which belongs to the Faculty of Agronomy and Biotechnology, China Agricultural College in Beijing, and Zhaoxian Experimental Station which belongs to the Institute of Grain and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences in Shijiazhuang, Hebei Province. Two therapies (NN&NP and LN&LP) had been carried out at Dishang Experimental Station (2024). The NN&NP remedy had 18 g N/m2 and eight g P/m2 utilized previous to sowing and 9.0 g N/m2 utilized on the stem elongation stage. The LN&LP remedy had no N and P utility. Twenty-five seeds of every line had been sown in 2 m rows, spaced 20 cm aside. Two consecutive discipline experiments had been additionally performed in Shangzhuang Experimental Station (2023–2024) with 15 seeds of every line sown in 1 m rows, spaced 20 cm aside. All rice crops had been grown beneath pure paddy circumstances on the Shangzhuang Experimental Station (2023). Seeds had been firstly sown within the seedbed for germination. After 21 days, seedlings grown within the seedbed had been transplanted into the paddy discipline with plant spacing: 15 cm, and row spacing: 25 cm.
Development circumstances and strategies of hydroponics experiment
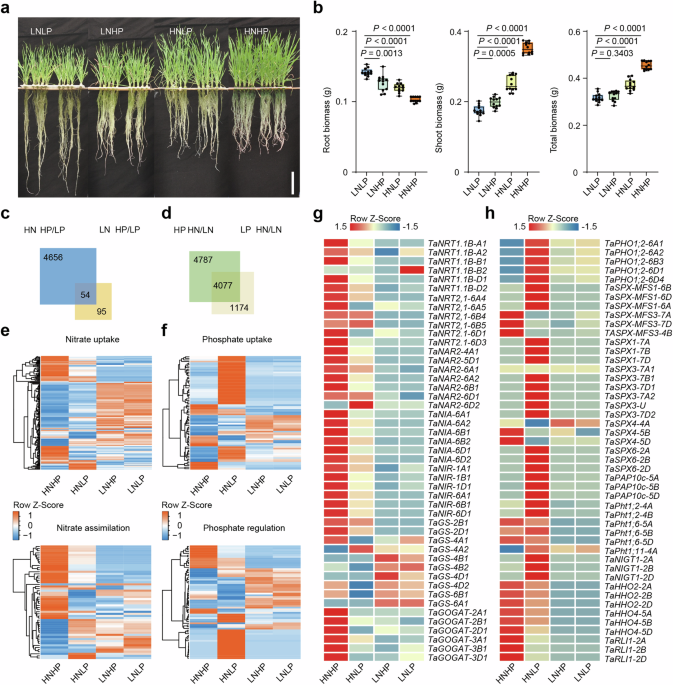
The wheat seeds (Fielder genetic background) had been initially germinated on moist filter papers for about 2 days. Then the seedlings had been cultivated within the plant hydroponic field (127 × 87 × 114 mm) for 4 days with 1 L distilled water, altering the water each 2 days. After elimination of residual endosperm, the seedlings had been transferred to plastic containers (30 × 15 × 15 cm) that contained 6 L of indicated nutrient resolution: HNHP (2.5 mM KNO3, 0.3 mM NaH2PO4), HNLP (2.5 mM KNO3, 0.03 mM NaH2PO4), LNHP (0.25 mM KNO3, 0.3 mM NaH2PO4), LNLP (0.25 mM KNO3, 0.03 mM NaH2PO4). The nutrient resolution was adjusted primarily based on basal nutrient resolution consists of the next macronutrients: 2.5 mM KNO3, 0.3 mM NaH2PO4·2H2O, 1 mM CaCl2·2H2O, 1 mM MgSO4·7H2O, 0.77 µM ZnSO4·7H2O, 0.32 µM CuSO4·5H2O, 40 µM EDTA–FeNa·3H2O, 9 µM MnSO4·4H2O, 0.05 µM (NH4)6·Mo7O24·4H2O and 20 µM H3BO3, with the pH adjusted to five.8. KNO3 and NaH2PO4·2H2O was added to the basal nutrient resolution at indicated concentrations for various therapies. The nutrient resolution was renewed each 3 days. 5 weeks after cultivation, wheat crops had been harvested for phenotypic analysis. All crops had been grown in plant development chambers (BPC500HL, Fujian Jiupo Biotechnology Co., Ltd) beneath 16 h of sunshine at 24 °C and eight h of darkish at 20 °C with about 70% relative humidity.
Assay for short-term nitrate-induced wheat transcriptome sequencing
For all of the short-term nitrate induction assays on this research, germinated seedlings (Chinese language spring) had been first grown in distilled water for two days. After elimination of residual endosperm, the seedlings had been then subjected to NO3− deficiency nutrient resolution (0.25 mM (NH4)2SO4 and a couple of.5 mM KCl) for 12 days. Subsequently, the seedlings had been uncovered to a nutrient resolution with 2.5 mM NO3− was added (0.25 mM (NH4)2SO4 and a couple of.5 mM KNO3−) for various durations. Whole RNA from entire crops was extracted utilizing the TRIzol reagent (Invitrogen), following the producer’s protocol.
Whole RNA extraction and actual time-PCR (RT-PCR)
Underneath regular circumstances and remedy, the roots of 14-day-old seedlings had been collected for RNA extraction. Whole RNAs had been extracted utilizing TRIzolTM Reagent (15596018, Thermo Fisher Scientific). The reverse transcription of cDNAs was carried out by utilizing HiScript II Q Choose RT SuperMix for qPCR (+gDNA wiper) (R223-01, Vazyme Biotech) in keeping with the producer’s suggestions. Taq Professional Common SYBR qPCR Grasp Combine (Q712-02, Vazyme Biotech) was used for RT-PCR evaluation. All primers had been proven in Supplementary Knowledge 12 and β-ACTIN was used as an inner management.
RNA-seq evaluation
Uncooked sequencing reads had been processed and filtered utilizing the software program fastp with the default parameters. The remaining high-quality reads had been mapped to the wheat IWGSC RefSeq v1.1 reference genome utilizing STAR with the default parameters. Characteristic Counts had been then used to summarize the reads mapped to every gene. To keep away from confounding results from low-quality gene fashions, solely high-confidence gene fashions with an FPKM of greater than 1 in at the very least one pattern had been thought of expressed. DESeq2 was used for differential expression evaluation. The genes with an absolute fold change worth larger than 1 and an FDR lower than 0.05 had been thought of DEGs. The GO enrichment evaluation was carried out utilizing GO Evaluation Toolkit and Datadase for Agriculture Neighborhood (agriGO), and phrases with an FDR lower than 0.05 had been thought of vital.
CUT&Tag assay
The CUT&Tag assays had been performed by Igenebook Biotechnology Co., Ltd in keeping with the beforehand described44. Wheat seedlings had been cleaved into nucleus suspension, and 10 w nuclei had been harvested and incubated with concanavalin A coated magnetic beads for 15 min at RT. Then, bead-bound cells had been resuspended and incubated with the suitable main antibody (1:50; F1804, Sigma) and IGG(1:50; 2729S, CST) in a single day at 4 °C. DNA from CUT&Tag was used to assemble sequencing libraries following the protocol supplied with Hyperactive Common CUT&Tag Assay Equipment for Illumina Professional (TD904, Vazyme Biotech Co., Ltd).
Evaluation of CUT&Tag information
Trimmomatic (model 0.36) was used to filter out low-quality reads. Clear reads had been mapped to the Chinese language Spring reference genome (IWGSC, RefSeq v1.1) by Bwa (model 0.7.15). Samtools (model 1.3.1) was used to take away potential PCR duplicates.MACS2 software program (model 2.1.1.20160309) was used to name peaks by default parameters (bandwidth, 300 bp; mannequin fold, 5, 50; p worth, 0.00001). If the midpoint of a peak is situated closest to the TSS of 1 gene, the height shall be assigned to that gene. HOMER (model 3) was used to foretell motif incidence inside peaks with default settings for a most motif size of 12 base pairs. ClusterProfiler (http://www.bioconductor.org/packages/launch/ bioc/html/clusterProfiler.html) within the R package deal was employed to carry out GO (http://geneontology.org/). The GO and KEGG enrichment evaluation had been calculated utilizing a hypergeometric distribution with a q worth cutoff of 0.05.
Yeast one-hybrid (Y1H) assay
The partial TaNRT2.1-6B promoter sequence was amplified, recombined with the pAbAi vector, after which built-in with the genome of the Y1HGold yeast pressure. CDS of the TaTCP6 was amplified, cloned into the pGADT7 vector, and transferred into the bait yeast pressure. The detailed process was in keeping with the Matchmaker Gold Y1H Library Screening System (Clontech). Totally different concentrations of the aureobasidin A (AbA) antibiotic had been used to pick out the constructive bait yeast pressure on a medium missing Ura and Leu (-L/-U).
Yeast two-hybrid (Y2H) assay
The coding sequence for the truncated TaTCP6 was amplified and cloned into the pGBKT7 vector to generate pGBKT7-TaTCP6tcp, which was used as a bait to display a yeast cDNA library derived from Fielder wheat seedlings to establish the candidate TaTCP6-interacting proteins. The Y2H screening was carried out in keeping with the consumer’s handbook (PT3024-1, Clontech Laboratories, Inc.) The coding sequences of genes of curiosity had been cloned into the pGADT7 vectors (Clontech), and varied combos of plasmids had been co-transformed into the yeast AH109 pressure. Constructive transformants had been chosen on artificial drop-out (SD) medium missing Trp and Leu (-L/-T), whereas the screening of interactions was carried out on SD medium missing Trp, Leu, His, and Ade (-L/-T/-H/-A).
Cut up-luciferase complementation imaging (LCI) assay
The CDSs of indicated genes had been individually cloned into the pCambia1300-nLUC (nLUC) and pCambia1300-cLUC (cLUC) vectors. The ensuing constructs had been remodeled into Agrobacterium tumefaciens pressure GV3101, which had been additional co-infiltrated in N. benthamiana leaves. About 36 h after infiltration, the LUC sign throughout the infiltration area was analyzed utilizing NightSHADE LB 985 (Berthold Applied sciences).
Bimolecular fluorescent complimentary (BiFC) assay
The CDSs of indicated genes had been individually cloned into the pUC57-cYFP and pUC57-nYFP vectors. The indicated constructs had been transiently expressed in wheat protoplasts utilizing a PEG-calcium mediated methodology as described beforehand45. The fluorescence indicators had been detected at 488 nm for YFP utilizing the confocal 507 microscope (LSM900; Carl Zeiss, Heidenheim, Germany).
Immunoprecipitation and co-immunoprecipitation assay
Whole proteins of wheat seedlings had been extracted with SDS-loading buffer (200 mM Tris pH 6.8, 40% glycerol, 8% SDS, and 20% β-mercaptoethanol) at 100 °C for five min. The Extracted proteins had been then separated on SDS-PAGE and transferred onto the PVDF membrane. The membrane-carrying proteins had been blocked with 5% milk in TBST containing 0.1% Tween-20 at room temperature for 1 h and incubated with the corresponding antibodies, corresponding to anti-rabbit IgG-HRP and anti-mouse IgG-HRP (Sigma-Aldrich). The proteins had been detected by IMMOBILON WESTERN CHEMILUM HRP SUBSTRATE (WBKLS, Millipore) beneath a fluorescence chemiluminescence imaging system (ChemiScope 6100). For Co-IP assays in N. benthamiana leaf tissues, the full-length CDS of indicated genes had been cloned into pCAMBIA1300-221-Flag, pSuper1300-MYC, pSuper1300-mCherry, and pSuper1300-GFP vector after which they had been remodeled into Agrobacterium strains and co-infiltrated into N. benthamiana leaves. About 36 h after infiltration, the overall proteins of infiltrated N. benthamiana leaves had been extracted with the lysis buffer as described above. For Co-IP assays in wheat protoplast, the full-length CDS of indicated genes had been cloned into pUC57-GFP, pUC57-MYC, and pUC57-Flag. About 16 h after infiltration, the overall proteins of the infiltrated wheat protoplast had been extracted with the lysis buffer as described above. Anti-GFP Magnetic Beads (M8823, Millipore) had been used for TaTCP6-GFP protein immunoprecipitation. The immunoprecipitated proteins had been detected by anti-Flag (1:5000; F3165, Sigma-Aldrich), anti-MYC (1:5000; ab9106, Abcam), anti-mCherry (1:2000; AE002, Abconal), anti-Actin (1:2000; AC009, Abconal) and anti-GFP antibodies (1:5000; ab32146, Abcam).
Subcellular localization
The CDS of TaTCP6, TaPHR2, TaRLI1, TaSPX1, TaSPX3, TaSPX4, and TaSPX6 was cloned into the pSuper1300-GFP vector and TaSPX4 was additionally cloned into the pSuper1300-mCherry vector. The indicated constructs had been transiently expressed in wheat protoplasts utilizing a PEG-calcium mediated methodology as described beforehand45. The fluorescence indicators had been detected at 488 nm for GFP and 546 nm for mCherry utilizing the confocal microscope (LSM900; Carl Zeiss, Heidenheim, Germany). The ensuing vectors of TaSPX1-GFP and TaSPX4-GFP had been additionally launched into Agrobacterium tumefaciens pressure GV3101 and the bacterial suspensions infiltrated into about 4-week-old N. benthamiana leaves. After 2 days, the infiltrated leaves had been collected and imaged on an LSM900 confocal microscope (Zeiss, Heidenheim, Germany). All primers used are listed in Supplementary Knowledge 12.
Twin-luciferase transcriptional exercise assay
The transcriptional actions of TaTCP6 on its downstream genes had been carried out in Nicotiana benthamiana. TaTCP6 had been cloned into the pSuper1300 vector as effectors. The promoter of TaNRT1.1B, TaNRT2.1, TaNIA1, TaNIR, TaIPS1.1, TaPht1;2, TaPht1;6, TaIPS1;2, TaPht1;11, and TaRLI1 was cloned into the dual-luciferase vector pGreenII 0800-LUC, respectively, to drive the firefly luciferase reporter gene (LUC), and luciferase from Renilla reniformis (REN) as an inner management was pushed by 35S promoter. All plasmids had been remodeled into Agrobacterium GV3101 and expressed in N. benthamiana leaves by Agrobacterium-mediated transient transformation. Contaminated tissues had been analyzed 48 h after infiltration. Actions of LUC and REN had been examined utilizing Twin-Luciferase Reporter Assay System equipment (E1960; Promega). The expression degree of a examined promoter is normalized by calculating the LUC:REN ratio.
Transient dual-luciferase assays had been additionally carried out to detect the impact of TaTCPs, TaSPX1, and TaTCPs-TaSPX1 advanced on the transcriptional activation exercise of TaPHR2. The TaPHR2 CDS had been cloned right into a vector to fuse TaPHR2 with the GAL4-BD area and generate the effector assemble BD-TaPHR2. TaTCPs and TaSPX1 had been additionally cloned into the pCAMBIA1300 vector respectively as effectors. To create the reporter assemble, a promoter with a 6× GAL4 upstream activating sequence and a TATA field was launched into the pGreenII 0800-LUC vector. The reporter and effector constructs had been co-transformed N. benthamiana leaves as indicated by Agrobacterium-mediated transient expression, utilizing the empty effector assemble GAL4-BD as a management. Contaminated tissues had been analyzed 48 h after infiltration. Actions of LUC and REN had been examined utilizing Twin-Luciferase Reporter Assay System equipment (E1960; Promega). The expression degree of a examined promoter is normalized by calculating the LUC:REN ratio.
Electrophoretic mobility shift assay (EMSA)
The total-length CDS of TaTCP6 had been cloned into pMAL-c2x vector to generate pMAL-c2x-TaTCP6. pMAL-c2x, and pMAL-c2x-TaTCP6 had been expressed within the E. coli Rosetta (DE3) pressure (Weidi, Shanghai, China) and purified utilizing amylose resin (New England Biolabs, Beijing, China) respectively. Oligonucleotide probes had been synthesized and their 5′ ends had been labeled with biotin. EMSA was carried out utilizing the Mild Shift Chemiluminescent EMSA Equipment (20148; Thermo Fisher Scientific), following the producer’s directions. The binding reactions had been carried out in keeping with the corresponding producer’s protocol. For the binding system of MBP-TaTCP6 fusion protein, about 2 μg protein, 0.02 μM biotin-labeled probe and acceptable quantity of aggressive probe had been incubated in 20 μL response mixtures containing 15 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM EDTA, 0.05% NP-40, 2 μg/μL BSA, 200 μM ZnCl2, 50 ng/μL poly (dI-dC), 5% glycerol for 20 min on 25 °C after which employed for electrophoretic evaluation.
Pull-down assay
MBP, MBP-TaTCP6, MBP-TaPHR2, and MBP-TaRLI1 fused proteins had been expressed within the E. coli Rosetta (DE3) pressure (Weidi, Shanghai, China) and purified utilizing amylose resin (New England Biolabs, Beijing, China) respectively. GST-TaSPX1 fused proteins had been expressed within the E. coli Rosetta (DE3) pressure (Weidi, Shanghai, China) and purified utilizing ProteinIso® GST Resin (Transgen Biotech, Beijing, China). HIS-TF-TaTCP6 For HIS fused protein was expressed within the E. coli Rosetta (DE3) pressure (Weidi, Shanghai, China) and purified utilizing ProteinIso® HIS Resin (Transgen Biotech, Beijing, China). pull-down assays, the GST-TaSPX1 protein was co-incubated individually with MBP, MBP-TaTCP6, MBP-TaPHR2, and MBP-TaRLI1 in lysis buffer (25 mM Tris (PH 7.5), 50 mM NaCl, 1 mM DTT) with amylose resin at 4 °C for two h. Afterward, the beads had been washed completely, and proteins had been analyzed by western blot utilizing anti-GST (1:2000; AE001, ABclonal), anti-HIS (1:2000; AE003, ABclonal) and anti-MBP (1:2000; AE016, ABclonal) antibodies.
Phylogenetic evaluation
To carry out phylogenetic evaluation, protein sequences of TCP gene household, GARP gene household, and SPX gene household from completely different plant species had been downloaded on Ensembl Crops (http://crops.ensembl.org/index.html). Mega X46 was employed to assemble the phylogenetic tree with the applications as follows: first, all protein sequences had been aligned in Clustal W with default parameters; second, the Most Probability methodology was used to construct phylogenetic tree with the Poisson mannequin and 1000 replicates bootstrap; subsequent, the phylogenetic tree was coloured and beautified on the web site Interactive Tree of Life.
Histological evaluation
The collected samples of mature wheat grains had been mounted in an FAA resolution [10% (v/v) formaldehyde, 50% (v/v) alcohol, 5% (v/v) acetic acid, and 35% (v/v) water overnight at 4 °C, and then were embedded in paraffin, dehydrated and decolorized as described previously47. The samples were then cut into 4-µm thick cross sections using a Leica Ultracut rotary microtome (Leica Biosystems, Wetzlar, Germany), and stained with Periodic acid Schiff (PAS) or 1% sarranine and 0.5% fast green (G1031, Servicebio, Wuhan). Photographs were taken with a microscope imaging system (DS-U3, Nikon, Japan).
15N uptake rate analysis
For 15N uptake rate analysis, wheat seedlings were cultured in the hydroponic culture (supplemented with 2.5 mM KNO3, pH 5.8) for 2 weeks. The seedlings were then transferred into modified hydroponic culture containing 2.5 mM 15N-KNO3 for 3 h. The roots of the treated seedlings were washed in 0.1 mM CaSO4 for 2 min to remove the unabsorbed 15N-KNO3 on the surface of the roots. Shoots and roots of the treated seedlings were collected separately and dried at 65 °C. The samples were ground to a fine powder and used for 15N content measurement using an isotope ratio mass spectrometer equipped with an elemental analyzer (Thermo Finnigan Delta Plus XP; Flash EA 1112).
Measurements of Pi content in wheat plants
A frozen sample after fresh weight measurement was homogenized in 0.1 mL Pi extraction buffer (0.1 M NaCl, 1 mM EDTA (pH 8.0, 10 mM Tris-HCl (pH 8.0), 0.07‰ (w/v) 2-hydroxy-1-ethanethiol, and 1 mM PMSF), using an ice-cold mortar and pestle. The homogenate was then diluted 10 times with 0.9 mL 1% (w/v) acetic acid and placed on ice for 30 min. After centrifugation at 10,000 × g for 10 min at 4 °C, The supernatant was collected, diluted to an appropriate concentration, then mixed with a molybdenum blue reagent (0.4% (w/v) ammonium molybdate melted in 0.5 m H2SO4 (solution A) was mixed with 10% ascorbic acid (solution B; A:B = 6:1) in a 7:3 ratio and incubated for 30 min at 42 °C. The absorption values of the solution at 820 nm were determined using a microplate reader (Tecan Spark). A standard curve was calculated from varying concentrations of KH2PO4, then the Pi content was calculated according to the standard curve and dilution ratio.
Pi-depletion assay
For the Pi-depletion experiment, a group of three seedlings was transferred into a flask with 500 mL of depletion solution (a modified nutrient salt depletion solution containing 100 μM NaH2PO4, Kimura B). All flasks were kept on a shaking table at 130 rpm. A 500 mL volume of depletion solution was withdrawn at the indicated time, and the Pi concentration was measured.
Measurement of nitrate reductase activity
Leaves of wheat seedlings growing under four different conditions were used to measure NR activity, following the instruction manual of the NR Kit (Solarbio LIFE SCIENCES, Beijing, China; BC0080). 0.1 g leaf samples were extracted in 1 mL extraction solution and the mixture was centrifuged at 4000 × g for 10 min. The resulting supernatant was collected for further analysis. The absorbance at 340 nm was used for the calculation of NR activity, one unit (U) of NR activity was defined as the amount of 1 μmol NADH consumed per hour per gram of sample48.
Nitrate concentration measurements
The nitrate concentrations in leaves of wheat seedlings were measured by the salicylic acid (SA) concentrated sulfuric acid colorimetry method. Samples of 1 g fresh wheat leaves or roots were homogenized, and 10 mL sterile water was added, and then nitrate was extracted for 30 min in a boiling water bath. 0.1 mL extracted solution, 0.4 mL 5% SA-concentrated sulfuric acid (w/v) solution, and 9.5 mL 8% NaOH (w/v) solution were well mixed and OD410 was measured when the solution was cooled to 25 °C. The nitrate concentration of the extracted solution was calculated using a standard curve.
Statistics and reproducibility
Quantitative data, including three biological and at least three technical replicates, are presented in the form of mean ± SD. Means of two samples were compared using Student’s two-tailed t-tests. Analysis of variance (one-way ANOVA) was conducted using GraphPad Prism 9.0.0 software with default parameters. Significant differences were determined by one-way ANOVA or the Student’s t-test: *P P
Three biological replicates were performed, with similar results; the results from one replicate are shown in the figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.