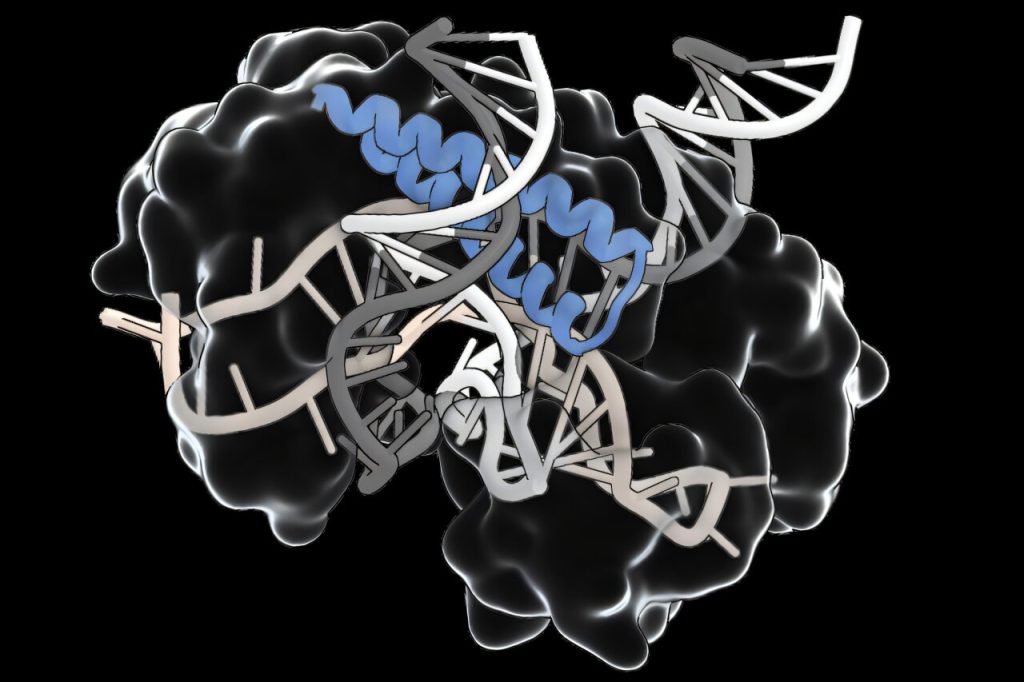

An enormous search of pure variety has led scientists at MIT’s McGovern Institute and the Broad Institute of MIT and Harvard to uncover historical methods with the potential to develop the genome-editing toolbox. These methods, which the researchers name TIGR (Tandem Interspaced Information RNA) methods, use RNA to information them to particular websites on DNA.
TIGR methods will be reprogrammed to focus on any DNA sequence of curiosity, they usually have distinct purposeful modules that may act on the focused DNA. Along with its modularity, TIGR could be very compact in comparison with different RNA-guided methods, like CRISPR, which is a significant benefit for delivering it in a therapeutic context.
These findings seem within the journal Science.
“It is a very versatile RNA-guided system with loads of various functionalities,” says Feng Zhang, the James and Patricia Poitras Professor of Neuroscience at MIT who led the analysis. The TIGR-associated (Tas) proteins that Zhang’s staff discovered share a attribute RNA-binding element that interacts with an RNA information that directs it to a particular website within the genome. Some minimize the DNA at that website, utilizing an adjoining DNA-cutting section of the protein. That modularity may facilitate instrument improvement, permitting researchers to swap helpful new options into pure Tas proteins.
“Nature is fairly unbelievable,” remarks Zhang, who can also be an investigator on the McGovern Institute and the Howard Hughes Medical Institute, a core member of the Broad Institute, a professor of mind and cognitive sciences and organic engineering at MIT, and co-director of the Ok. Lisa Yang and Hock E. Tan Heart for Molecular Therapeutics at MIT.
“It is acquired an amazing quantity of variety, and we’ve got been exploring that pure variety to seek out new organic mechanisms and harnessing them for various functions to govern organic processes,” he says.
Beforehand, Zhang’s staff had tailored bacterial CRISPR methods into gene-editing instruments which have reworked trendy biology. His staff has additionally discovered quite a lot of programmable proteins, each from CRISPR methods and past.
Of their new work, to seek out novel programmable methods, the staff started by zeroing in on a structural characteristic of the CRISPR Cas9 protein that binds to the enzyme’s RNA information. That may be a key characteristic that has made Cas9 such a strong instrument.
“Being RNA-guided makes it comparatively simple to reprogram, as a result of we all know how RNA binds to different DNA or different RNA,” Zhang explains. His staff searched a whole lot of thousands and thousands of organic proteins with recognized or predicted constructions, searching for any that shared an identical area. To search out extra distantly associated proteins, they used an iterative course of: from Cas9, they recognized a protein known as IS110, which had beforehand been proven by others to bind RNA. They then zeroed in on the structural options of IS110 that allow RNA binding and repeated their search.
At this level, the search had turned up so many distantly associated proteins that the staff turned to synthetic intelligence to make sense of the checklist.
“If you end up doing iterative, deep mining, the ensuing hits will be so various that they’re tough to research utilizing normal phylogenetic strategies, which depend on conserved sequences,” explains Guilhem Faure, a computational biologist in Zhang’s lab.
With a protein giant language mannequin, the staff was capable of cluster the proteins that they had discovered into teams in response to their probably evolutionary relationships. One group set other than the remainder, and its members have been notably intriguing as a result of they have been encoded by genes with often spaced repetitive sequences paying homage to a vital part of CRISPR methods. These have been the TIGR-Tas methods.
Zhang’s staff found over 20,000 completely different Tas proteins, principally occurring in bacteria-infecting viruses. Sequences inside every gene’s repetitive area—its TIGR arrays—encode an RNA information that interacts with the RNA-binding a part of the protein. In some, the RNA-binding area is adjoining to a DNA-cutting a part of the protein. Others seem to bind to different proteins, which suggests they may assist direct these proteins to DNA targets.
Zhang and his staff experimented with dozens of Tas proteins, demonstrating that some will be programmed to make focused cuts to DNA in human cells. As they consider growing TIGR-Tas methods into programmable instruments, the researchers are inspired by options that would make these instruments notably versatile and exact.
They word that CRISPR methods can solely be directed to segments of DNA which are flanked by brief motifs often called PAMs (protospacer adjoining motifs). TIGR Tas proteins, in distinction, haven’t any such requirement.
“This implies theoretically, any website within the genome ought to be targetable,” says scientific advisor Rhiannon Macrae.
The staff’s experiments additionally present that TIGR methods have what Faure calls a “dual-guide system,” interacting with each strands of the DNA double helix to dwelling in on their goal sequences, which ought to guarantee they act solely the place they’re directed by their RNA information. What’s extra, Tas proteins are compact—1 / 4 of the dimensions of Cas9 on common—making them simpler to ship, which may overcome a significant impediment to therapeutic deployment of gene modifying instruments.
Excited by their discovery, Zhang’s staff is now investigating the pure position of TIGR methods in viruses in addition to how they are often tailored for analysis or therapeutics. They’ve decided the molecular construction of one of many Tas proteins they discovered to work in human cells, and can use that info to information their efforts to make it extra environment friendly. Moreover, they word connections between TIGR-Tas methods and sure RNA-processing proteins in human cells.
“I believe there’s extra there to review by way of what a few of these relationships could also be, and it could assist us higher perceive how these methods are utilized in people,” Zhang says.
Extra info:
Guilhem Faure et al, TIGR-Tas: A household of modular RNA-guided DNA-targeting methods in prokaryotes and their viruses, Science (2025). DOI: 10.1126/science.adv9789
Offered by
McGovern Institute for Mind Analysis
Quotation:
An historical RNA-guided system may simplify supply of gene modifying therapies (2025, February 27)
retrieved 1 March 2025
from https://phys.org/information/2025-02-ancient-rna-delivery-gene-therapies.html
This doc is topic to copyright. Other than any honest dealing for the aim of personal examine or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.