
Antibiotic resistance is a world concern that threatens our capacity to forestall and deal with bacterial infections in people and animals. To higher monitor the emergence and unfold of resistance, researchers on the Carl R. Woese Institute for Genomic Biology have developed a CRISPR-enriched metagenomics technique for the improved surveillance of antibiotic resistance genes (ARGs) in wastewater.
The analysis is printed within the journal Water Analysis.
Whereas antibiotics are highly effective trendy instruments for combating infections, micro organism change and adapt over time in response to antibiotic publicity, subsequently reducing effectiveness. Widespread overuse and misuse of antibiotics within the well being care and meals industries additional speed up this downside.
Past direct publicity to antibiotics, resistance can also be handed between totally different micro organism by means of the switch of small items of bacterial DNA known as antibiotic resistance genes. There are over 5,000 recognized ARGs, and these genes will be present in scientific samples, in addition to our bodies of water, originating from hospitals, farms, and sewage methods.
“ARGs can cut back the life-saving energy of medicine used to deal with bacterial infections,” mentioned Helen Nguyen (IGOH), a professor of civil and environmental engineering on the College of Illinois Urbana-Champaign. “Wastewater detection of ARGs with scientific significance permits public well being authorities and physicians to anticipate what’s circulating in communities.”
Wastewater accommodates quite a few totally different ARGs blended along with genetic materials from varied sources, together with people, viruses, and micro organism. As a result of ARGs solely make up a tiny proportion of the overall DNA content material, uncovering them in wastewater samples requires delicate detection strategies. The commonest method is quantitative polymerase chain response (qPCR). This technique makes use of RNA guides known as primers to determine the precise DNA sequences of recognized ARGs, that are then amplified for detection.
“qPCR is a delicate technique that many individuals in public well being are well-trained to do, however it requires major design and validation, which may be very time-consuming,” mentioned Yuqing Mao, a doctoral pupil in civil and environmental engineering who was the primary writer of the paper. “Since qPCR is used to tug out focused gene sequences, all the opposite genetic materials within the pattern stays fully unknown.”
The second technique, metagenomics, isn’t as delicate as qPCR, however captures a extra full story of the genetic data contained in a pattern. Metagenomics entails breaking all of the pattern DNA into thousands and thousands of smaller fragments that are concurrently sequenced utilizing subsequent era sequencing applied sciences. Computational algorithms piece collectively the total DNA sequences for comparability in opposition to databases to find out their identities.
“ARGs make up lower than 1%—most likely even nearer to 0.1%—of DNA within the pattern. Utilizing normal metagenomics strategies, 99.9% of the DNA detected isn’t related to ARGs,” Mao mentioned.
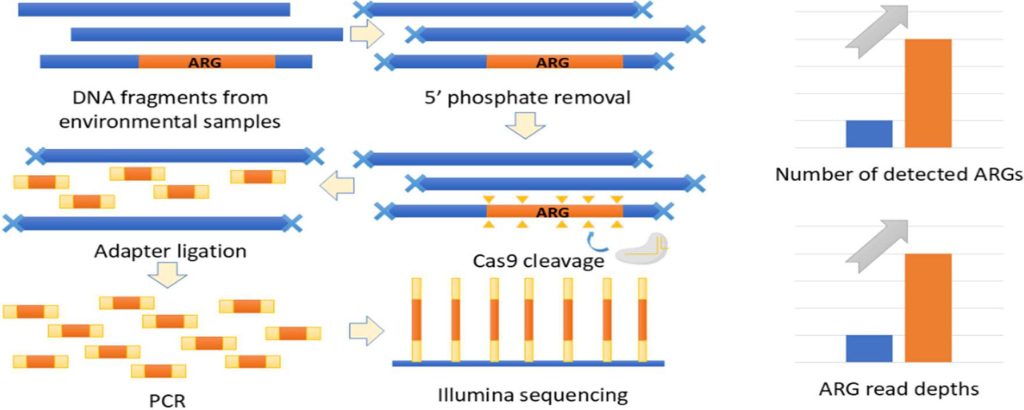
To complement the quantity of ARG-associated fragments within the samples, Mao, Nguyen, and their collaborator, Joanna Shisler, who’s affiliated with the Division of Microbiology at Illinois, leveraged the CRISPR-Cas9 system—a extremely efficient software for gene modifying.
The DNA is fragmented in random places when utilizing normal metagenomics strategies, however the incorporation of CRIPSR-Cas9 permits for focused fragmentation inside ARGs. By designing a pool of 6,010 totally different information RNAs that might particularly bind to DNA at totally different websites present in ARGs, the Cas9 protein might be directed to chop at these places.
“Our new CRISPR technique will increase the abundance of ARG fragments within the pattern, which will increase their probabilities of being learn and detected. CRISPR additionally has higher potential for multiplexed assays than one thing like PCR as a result of the molecular interplay is easy and simple for CRISPR,” Mao mentioned.
Their new technique lowered the detection restrict of ARGs by an order of magnitude, from 10-4 to 10-5, in comparison with normal metagenomics, and located 1,189 extra ARGs and 61 extra ARG households which are low in abundance in wastewater samples.
As a sixth-year graduate pupil, Mao constructed this venture from the bottom up—overcoming scientific obstacles and studying many new methods alongside the best way. She mentioned, “The primary time I acquired the sequencing outcomes, I by no means anticipated how rather more delicate it could be in comparison with the common technique—it detected many extra ARGs than we thought it could. After ending the venture, I really feel like I’ve grown up.”
However whereas this work is wrapped up, Mao and Nguyen are already pursuing a number of new instructions, together with increasing the functions of their CRISPR-Cas9 metagenomic technique to a broader vary of environmental samples and utilizing their outcomes to information the design of recent qPCR primers.
Extra data:
Yuqing Mao et al, Enhanced detection for antibiotic resistance genes in wastewater samples utilizing a CRISPR-enriched metagenomic technique, Water Analysis (2024). DOI: 10.1016/j.watres.2024.123056
Offered by
College of Illinois at Urbana-Champaign
Quotation:
CRISPR-based technique enhances detection of antibiotic resistance in wastewater (2025, March 3)
retrieved 4 March 2025
from https://phys.org/information/2025-03-crispr-based-method-antibiotic-resistance.html
This doc is topic to copyright. Aside from any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is offered for data functions solely.