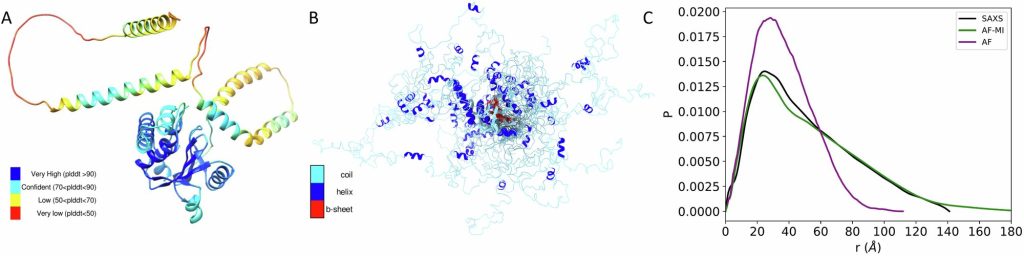

Intrinsically disordered proteins (IDPs) don’t attain a steady secondary or tertiary construction and quickly change their conformation, making construction prediction significantly difficult. Though these proteins exhibit chaotic and “disordered” buildings, they nonetheless carry out important features.
IDPs comprise roughly 30% of the human proteome and play essential useful roles in transcription, translation, and signaling. Many mutations linked to neurological ailments, together with amyotrophic lateral sclerosis (ALS), are positioned in intrinsically disordered protein areas (IDRs).
Highly effective machine-learning algorithms, together with AlphaFold and RoseTTAFold, can not present sensible representations of those ‘disordered’ and ‘chaotic’ protein areas as a complete. It is because they haven’t been skilled on such knowledge and since these proteins exhibit inherent dynamic conduct, adopting a variety of conformations quite than a single steady one.
Now, a crew of researchers from BSRC Fleming and the Centre for Misfolding Ailments on the College of Cambridge has discovered an environment friendly approach to predict the buildings of a big fraction of all human proteins that had been beforehand thought of “darkish” and notoriously tough to watch.
The crew developed and used an algorithm known as “AlphaFold-Metainference,” which was skilled on knowledge from accessible protein construction databases in addition to molecular dynamics simulations. The findings of the research had been just lately revealed in Nature Communications.

“AlphaFold has reworked structural biology by offering correct predictions of protein buildings. We’ve got now proven find out how to prolong these predictions to IDPs, which make up a few third of the human proteome and are implicated in just about all main ailments,” says Michele Vendruscolo, Professor of Biophysics on the Centre for Misfolding Ailments on the College of Cambridge.
“We had been stunned to seek out that though AlphaFold doesn’t precisely predict the three-dimensional construction of IDPs, it could actually predict the distances between amino acids with fairly good accuracy. We then integrated this info into molecular dynamics simulations, permitting us to precisely predict the three-dimensional buildings these disordered proteins undertake and their movement,” explains Dr. Faidon Brotzakis, a senior postdoctoral researcher within the lab of Dr. Georgios Skretas on the Institute for Bioinnovation of the Biomedical Sciences Analysis Heart “Alexander Fleming” (BSRC Fleming) and the research’s first writer.
The algorithm was examined on proteins containing each disordered and non-disordered areas, together with TDP-43 (related to ALS), ataxin-3 (linked to Machado-Joseph illness), and the prion protein (implicated in Creutzfeldt-Jakob illness).
“We examined the algorithm on a complete of 11 IDPs and 6 PDPs, however we centered significantly on proteins related to severe ailments. In all circumstances, the algorithm outperformed AlphaFold in accuracy. Actually, in 80% of circumstances, it matched or exceeded the accuracy of molecular dynamics simulations. This demonstrates the algorithm’s benefit within the structural characterization of IDPs,” explains Dr. Brotzakis.
Scientists now have a sooner and extra correct approach to decide the buildings of disordered proteins, particularly in circumstances the place experimental knowledge are unavailable. “Sooner or later, we will use this info to find molecules of pharmaceutical curiosity that may work together strongly with these proteins and modify their dynamics. This might forestall their problematic folding into poisonous types, similar to amyloid fibrils, that are noticed in lots of neurodegenerative ailments,” says Brotzakis.
The subsequent steps are to use the algorithm to different biomolecules, similar to DNA and RNA.
Extra info:
Z. Faidon Brotzakis et al, AlphaFold prediction of structural ensembles of disordered proteins, Nature Communications (2025). DOI: 10.1038/s41467-025-56572-9
Offered by
Biomedical Sciences Analysis Heart Alexander Fleming
Quotation:
Algorithm sheds mild on ‘disordered’ proteins as soon as thought of too tough to check (2025, March 6)
retrieved 6 March 2025
from https://phys.org/information/2025-03-algorithm-disordered-proteins-difficult.html
This doc is topic to copyright. Other than any honest dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is offered for info functions solely.