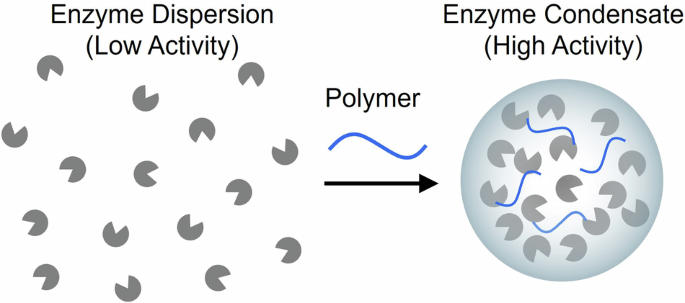
van Beilen JB, Li Z. Enzyme expertise: an outline. Curr Opin Biotechnol. 2002;13:338–44.
Eijsink VGH, Bjørk A, Gåseidnes S, Sirevåg R, Synstad B, van den Burg B, et al. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–20.
Eijsink VGH, Gåseidnes S, Borchert TV, van den Burg B. Directed evolution of enzyme stability. Biomol Eng. 2005;22:21–30.
H Stamatis, Multienzymatic assemblies: Strategies and protocols, 1st ed., Springer, New York, NY, 2022.
Schmitt DL, An S. Spatial group of metabolic enzyme complexes in cells. Biochemistry. 2017;56:3184–96.
Miura N, Shinohara M, Tatsukami Y, Sato Y, Morisaka H, Kuroda Okay, et al. Spatial reorganization of Saccharomyces cerevisiae enolase to change carbon metabolism beneath hypoxia. Eukaryot Cell. 2013;12:1106–19.
Jin M, Fuller GG, Han T, Yao Y, Alessi AF, Freeberg MA, et al. Glycolytic enzymes coalesce in G our bodies beneath hypoxic stress. Cell Rep. 2017;20:895–908.
An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in residing cells. Science. 2008;320:103–6.
Pedley AM, Benkovic SJ. A brand new view into the regulation of purine metabolism: the purinosome. Traits Biochem Sci. 2017;42:141–54.
Lim S, Clark DS. Part-separated biomolecular condensates for biocatalysis. Traits Biotechnol. 2024;42:496–509.
Yewdall NA, André AAM, Lu T, Spruijt E. Coacervates as fashions of membraneless organelles. Curr Opin Colloid Interface Sci. 2021;52:101416.
Nakashima KK, André AAM, Spruijt E. Enzymatic management over coacervation. Strategies Enzymol. 2021;646:353–89.
Horn JM, Kapelner RA, Obermeyer AC. Macro- and microphase separated protein-polyelectrolyte complexes: Design parameters and present progress. Polymers. 2019;11:578.
Ura T, Kagawa A, Sakakibara N, Yagi H, Tochio N, Kigawa T, et al. T. Mikawa, Activation of L-lactate oxidase by the formation of enzyme assemblies via liquid–liquid section separation. Sci Rep. 2023;13:1–9.
Ura T, Sakakibara N, Hirano Y, Tamada T, Takakusagi Y, Shiraki Okay. T. Mikawa, Activation of oxidoreductases by the formation of enzyme meeting. Sci Rep. 2023;13:1–8.
Cummings CS, Obermeyer AC. Part separation habits of supercharged proteins and polyelectrolytes. Biochemistry. 2018;57:314–23.
Koga S, Williams DS, Perriman AW, Mann S. Peptide-nucleotide microdroplets as a step in direction of a membrane-free protocell mannequin. Nat Chem. 2011;3:720–4.
Baksi A, Zerze H, Agrawal A, Karim A, Zerze GH. The molecular image of the native atmosphere in a steady mannequin coacervate. Commun Chem. 2024;7:222.
Kota D, Prasad R, Zhou H-X. Adenosine triphosphate mediates section separation of disordered fundamental proteins by bridging intermolecular interplay networks. J Am Chem Soc. 2024;146:1326–36.
Ura T, Tomita S, Shiraki Okay. Dynamic habits of liquid droplets with enzyme compartmentalization triggered by sequential glycolytic enzyme reactions. Chem Commun. 2021;57:12544–7.
Jaworek MW, Oliva R, Winter R. Enabling excessive activation of glucose-6-phosphate dehydrogenase exercise via liquid condensate formation and compression, Chem. Eur. J. 2024;30:e202400690.
Blocher McTigue WC, Perry SL. Chapter Ten – Incorporation of proteins into advanced coacervates, in: CD Keating (Ed.), Strategies in Enzymology, Educational Press, 2021: pp. 277–306.
Blocher McTigue WC, Perry SL. Protein encapsulation utilizing advanced coacervates: What nature has to show us. Small. 2020;16:e1907671.
Sakakibara N, Ura T, Mikawa T, Sugai H, Shiraki Okay. Transient formation of multi-phase droplets brought on by the addition of a folded protein into advanced coacervates with an oppositely charged floor relative to the protein. Comfortable Matter. 2023;19:4642–50.
Jurrus E, Engel D, Star Okay, Monson Okay, Brandi J, Felberg LE, et al. Enhancements to the APBS biomolecular solvation software program suite. Protein Sci. 2018;27:112–28.
Nomoto A, Shiraki Okay. Thermal aggregation of immunoglobulin G relying on the cost state of protein-polyelectrolyte complexes. Int J Biol Macromol. 2025;296:139500.
O’Flynn BG, Mittag T. The position of liquid-liquid section separation in regulating enzyme exercise. Curr Opin Cell Biol. 2021;69:70–79.
Dindo M, Bevilacqua A, Laurino P. Enzymes and liquid-liquid section separation: a brand new period for the regulation of enzymatic exercise. Seibutsu Butsuri Kagaku. 2023;63:12–15.
Zhang Y, Narlikar GJ, Kutateladze TG. Enzymatic Reactions inside Organic Condensates. J Mol Biol. 2021;433:166624.
Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, et al. The Eukaryotic CO2-concentrating organelle is liquid-like and displays dynamic reorganization. Cell. 2017;171:148–.e19.
Du M, Chen ZJ. DNA-induced liquid section condensation of cGAS prompts innate immune signaling. Science. 2018;361:704–9.
Peeples W, Rosen MK. Mechanistic dissection of elevated enzymatic charge in a phase-separated compartment. Nat Chem Biol. 2021;17:693–702.
Forman-Kay JD, Ditlev JA, Nosella ML, Lee HO. What are the distinguishing options and measurement necessities of biomolecular condensates and their implications for RNA-containing condensates?. RNA. 2022;28:36–47.
Kohnhorst CL, Kyoung M, Jeon M, Schmitt DL, Kennedy EL, Ramirez J, et al. Identification of a multienzyme advanced for glucose metabolism in residing cells. J Biol Chem. 2017;292:9191–203.
Gil-Garcia M, Benítez-Mateos AI, Papp M, Stoffel F, Morelli C, Normak Okay, et al. Native atmosphere in biomolecular condensates modulates enzymatic exercise throughout size scales. Nat Commun. 2024;15:3322.
Tibble RW, Gross JD. A name to order: Inspecting structured domains in biomolecular condensates. J Magn Reson. 2023;346:107318.
Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, et al. Part transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–47.
Lee M-Y, Dordick JS. Enzyme activation for nonaqueous media. Curr Opin Biotechnol. 2002;13:376–84.
Visser BS, Lipiński WP, Spruijt E. The position of biomolecular condensates in protein aggregation. Nat Rev Chem. 2024;8:686–700.
Dong H, Qin S, Zhou H-X. Results of macromolecular crowding on protein conformational modifications. PLoS Comput Biol. 2010;6:e1000833.
Abyzov A, Blackledge M, Zweckstetter M. Conformational dynamics of intrinsically disordered proteins regulate biomolecular condensate chemistry. Chem Rev. 2022;122:6719–48.
Saini B, Mukherjee TK. Biomolecular condensates regulate enzymatic exercise beneath a crowded milieu: Synchronization of liquid-liquid section separation and enzymatic transformation. J Phys Chem B. 2023;127:180–93.
Scholl D, Deniz AA. Conformational freedom and topological confinement of proteins in biomolecular condensates. J Mol Biol. 2022;434:167348.
Tibble RW, Depaix A, Kowalska J, Jemielity J, Gross JD. Biomolecular condensates amplify mRNA decapping by biasing enzyme conformation. Nat Chem Biol. 2021;17:615–23.
Kurinomaru T, Tomita S, Hagihara Y, Shiraki Okay. Enzyme hyperactivation system based mostly on a complementary charged pair of polyelectrolytes and substrates. Langmuir. 2014;30:3826–31.
J. Thiele M, Davari MD, König M, Hofmann I, Junker NO, Mirzaei Garakani T, et al. Enzyme–polyelectrolyte complexes enhance the catalytic efficiency of enzymes. ACS Catal. 2018;8:10876–87.
Ito-Harashima S, Miura N. Compartmentalization of a number of metabolic enzymes and their preparation in vitro and in cellulo. Biochim Biophys Acta Gen Subj. 2025;1869:130787.
Harris R, Berman N, Lampel A. Coacervates as enzymatic microreactors, Chem Soc Rev. 2025. https://doi.org/10.1039/d4cs01203h.
Wan L, Ke J, Zhu Y, Zhang W, Mu W. Latest advances in engineering artificial biomolecular condensates. Biotechnol Adv. 2024;77:108452.
Arnold FH. Design by directed evolution. Acc Chem Res. 1998;31:125–31.
Maghraby YR, El-Shabasy RM, Ibrahim AH, Azzazy HME-S. Enzyme immobilization applied sciences and industrial functions. ACS Omega. 2023;8:5184–96.
Garcia-Galan C, Berenguer-Murcia Á, Fernandez-Lafuente R, Rodrigues RC. Potential of various enzyme immobilization methods to enhance enzyme efficiency. Adv Synth Catal. 2011;353:2885–904.
Perumal S, Atchudan R, Lee W. A evaluate of polymeric micelles and their functions. Polymers. 2022;14:2510.
Harada A, Kataoka Okay. Novel polyion advanced micelles entrapping enzyme molecules within the core: Preparation of narrowly-distributed micelles from lysozyme and poly(ethylene glycol)−Poly(aspartic acid) block copolymer in aqueous medium. Macromolecules. 1998;31:288–94.
Solar J, Li Z. Polyion complexes by way of electrostatic interplay of oppositely charged block copolymers. Macromolecules. 2020;53:8737–40.
Wang H, Vant JW, Zhang A, Sanchez RG, Wu Y, Micou ML, et al. Group of a useful glycolytic metabolon on mitochondria for metabolic effectivity. Nat Metab. 2024;6:1712–35.
Zhou J, Liu C, Yu H, Tang N, Lei C. Analysis progresses and software of biofuel cells based mostly on immobilized enzymes. Appl Sci. 2023;13:5917.
Sassolas A, Blum LJ, Leca-Bouvier BD. Immobilization methods to develop enzymatic biosensors. Biotechnol Adv. 2012;30:489–511.
Nobeyama T, Yoshida T, Shiraki Okay. Interfacial and intrinsic molecular results on the section separation/transition of heteroprotein condensates. Int J Biol Macromol. 2024;254:128095.
Biplab KC, Nii T, Mori T, Katayama Y, Kishimura A. Dynamic annoyed cost hotspots created by cost density modulation sequester globular proteins into advanced coacervates. Chem Sci. 2023;14:6608–20.
Yoshida T, Sakakibara N, Ura T, Minamiki T, Shiraki Okay. Cationic polyelectrolytes forestall the aggregation of l-lactate dehydrogenase beneath unstable circumstances, Int J Biol Macromol. 2023;257:128549.
Voets IK, de Keizer A, Cohen Stuart MA. Complicated coacervate core micelles. Adv Colloid Interface Sci. 2009;147–148:300–18.
Wei M, Gao Y, Li X, Serpe MJ. Stimuli-responsive polymers and their functions. Polym Chem. 2017;8:127–43.
Panganiban B, Qiao B, Jiang T, DelRe C, Obadia MM, Nguyen TD, et al. Random heteropolymers protect protein operate in overseas environments. Science. 2018;359:1239–43.
Waltmann C, Mills CE, Wang J, Qiao B, Torkelson JM, Tullman-Ercek D, et al. Practical enzyme-polymer complexes. Proc Natl Acad Sci Usa. 2022;119:e2119509119.
Macazo FC, Minteer SD. Enzyme cascades in biofuel cells. Curr Opin Electrochem. 2017;5:114–20.
Lopez-Gallego F, Schmidt-Dannert C. Multi-enzymatic synthesis. Curr Opin Chem Biol. 2010;14:174–83.
Bowie JU, Sherkhanov S, Korman TP, Valliere MA, Opgenorth PH, Liu H. Artificial biochemistry: The bio-inspired cell-free method to commodity chemical manufacturing. Traits Biotechnol. 2020;38:766–78.
Liu M, He S, Cheng L, Qu J, Xia J. Part-separated multienzyme biosynthesis. Biomacromolecules. 2020;21:2391–9.
Nakashima KK, Baaij JF, Spruijt E. Reversible era of coacervate droplets in an enzymatic community. Comfortable Matter. 2018;14:361–7.
Miura N. Condensate Formation by Metabolic Enzymes in Saccharomyces cerevisiae. Microorganisms. 2022;10:232.
Kluczka E, Rinaldo V, Coutable-Pennarun A, Stines-Chaumeil C, Anderson JLR, Martin N. Enhanced catalytic exercise of a de novo enzyme in a coacervate section. ChemCatChem. 2024;16:e202400558.