Evaluating medical NLP device outputs
All phenotype information utilized in our preliminary publications12,13 was generated from affected person clinic notes utilizing the CNLP software program CLiX. CLiX is a proprietary medical NLP expertise developed by the business healthcare analytics firm Clinithink18. Given the final word purpose of creating MPSE for adoption by various hospital and clinic techniques, we sought to find out MPSE’s efficiency utilizing phenotype information produced by different instruments as nicely. We carried out a collection of analyses on 5 completely different CNLP instruments to check their relative utility to be used with MPSE: ClinPhen19, CLiX, cTAKES20, MedLEE21, and MetaMapLite22. A quick description of those instruments is given in Supplementary Desk 1. Earlier than assessing MPSE’s efficiency utilizing phenotype information produced by these completely different CNLP instruments, we first in contrast the phenotype descriptions (HPO time period units) generated by these instruments utilizing the identical units of clinic notes—on this case, the notes from 1838 College of Utah NICU admits.
Time period counts
Abstract statistics for distinctive HPO time period counts generated by every CNLP device in addition to the “handbook” time period units recognized by skilled physicians are given in Supplementary Desk 2. The HPO time period units used all through this work had been pre-processed by eradicating guardian phrases to maintain solely essentially the most particular phenotype phrases. In each case, the CNLP instruments all produced bigger HPO time period units per affected person than did skilled assessment. Amongst HPO time period units for the College of Utah NeoSeq sufferers, MedLEE yielded the fewest phrases (common 31.4 phrases per affected person) whereas CLiX yielded essentially the most phrases (common 111.2 phrases per affected person), almost twice as many as the following most prolific device cTAKES (70.5 phrases per affected person). Unsequenced College of Utah NICU sufferers had considerably fewer phrases of their phenotype descriptions than NeoSeq sufferers, in keeping with our observations from different affected person cohorts12.
Semantic similarity
We additionally calculated pairwise semantic similarity coefficients throughout all of the NeoSeq phenotype units. Semantic similarity is completely different from strict identity-based similarity measures, akin to unweighted Jaccard similarity, in that two phrases could be non-identical however nonetheless contribute positively to the similarity coefficient if they’re neighbors, i.e., they lie close to each other within the HPO directed acyclic graph. To estimate the chance that the semantic similarities of the time period units produced by the instruments are statistically completely different from a null or random distribution, for every pairwise comparability, we sampled the HPO to yield two random time period units with sizes equivalent to the unique units. Semantic similarity coefficients had been then calculated for these randomized units and plotted alongside the true information in Supplementary Fig. 1. The pronounced separation between semantic similarity distributions of actual and simulated information suggests the HPO time period units produced by the instruments displays a standard underlying phenotypic actuality that’s being recognized to a better or lesser diploma by all of the instruments. In keeping with this, the distinction in imply similarity between actual and simulated datasets is very statistically vital for each device by Scholar’s paired T-test, even after a number of check corrections (information not proven).
NLP sensitivity and accuracy
Subsequent, we carried out an orthogonal check of CNLP device sensitivity and accuracy by calculating the overlap between the CNLP-generated phenotype units and two distinct “floor reality” reference units: expert-generated phenotypes and OMIM disease-associated phenotypes. The skilled reference units constituted the HPO time period lists manually curated by physicians for the 65 sequenced College of Utah NICU sufferers23. The OMIM illness reference units had been restricted to the HPO phenotype phrases related to the OMIM illness prognosis for the subset of 26 WGS-diagnosed Utah NICU sufferers. For these analyses, a CNLP time period is taken into account a “true optimistic” if it or any of its guardian phrases are discovered within the floor reality set. This strategy is justified by the ontological relationship between guardian and little one phrases in HPO, the place a toddler time period inherently implies the presence of its guardian time period(s). Since HPO is structured in a fashion the place extra particular phrases (i.e., little one phrases) characterize refined phenotypic descriptions, they subsume the extra normal phrases (i.e., guardian phrases). For instance, having Thrombocytosis (HP:0001894) essentially signifies the presence of the guardian phenotype Irregular platelet rely (HP:0011873).
Supplementary Fig. 2 reveals sensitivity and accuracy distributions for CNLP phrases units in contrast with doctor handbook phrases and OMIM disease-associated phrases. The relative sensitivity among the many CNLP instruments roughly correlates with the instruments’ common time period counts (see Supplementary Desk 2), which isn’t stunning; the extra phrases a device generates, the extra seemingly it is going to seize phrases within the reference set. CLiX had the best common sensitivity among the many instruments (handbook phrases sensitivity: 63%; OMIM phrases sensitivity: 21%), whereas MetaMapLite had the bottom (handbook phrases sensitivity: 15%; OMIM phrases sensitivity: 9%). The accuracy measure differs from sensitivity by controlling for the variable sizes of the CNLP time period units. Thus, a CNLP device with excessive sensitivity might have a low relative accuracy if it has many extra phrases than one other device with decrease sensitivity. ClinPhen had the best common accuracy among the many instruments (handbook phrases sensitivity: 13%; OMIM phrases sensitivity: 11%), whereas MetaMapLite had the bottom (handbook phrases sensitivity: 2%; OMIM phrases sensitivity: 4%). Regardless of the modest sensitivity and accuracy of those CNLP instruments in comparison with the “floor reality” units, MPSE and different phenotype-driven medical diagnostics instruments akin to GEM11, seem like very strong towards “noisy” phenotype information inputs (see subsequent outcomes for justification).
MPSE flexibly handles enter information from a wide range of sources
To additional consider the sensible utility of the MPSE algorithm, we carried out a comparative evaluation of various CNLP instruments and information sorts as inputs to MPSE. This evaluation addresses a number of important issues for the deployment of MPSE in various medical environments. First, we explored the interoperability of MPSE by assessing whether or not a mannequin educated with information from one CNLP device might reliably predict outcomes utilizing information generated by a distinct device. Second, we investigated the feasibility of utilizing non-phenotype information with MPSE. Given the final word purpose of creating MPSE for adoption by various hospital and clinic techniques, the overarching goal of those analyses is to raised perceive MPSE’s flexibility, robustness, and broader applicability in real-world medical settings.
MPSE efficiency utilizing completely different CNLP instruments
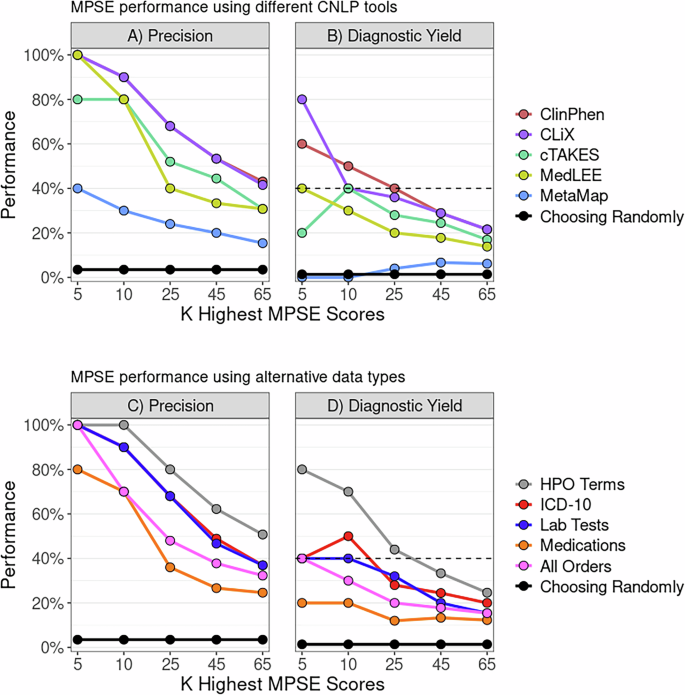
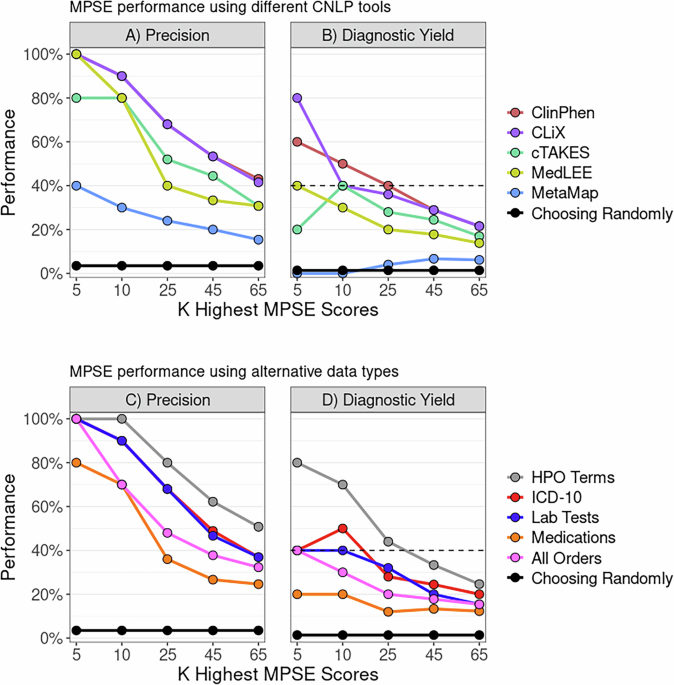
To find out whether or not MPSE, educated with information from one CNLP device, can reliably predict outcomes utilizing information generated by a distinct CNLP device, we started with the unique MPSE mannequin educated utilizing CLiX-generated phenotype information from the RCHSD cohort. This mannequin was used to make predictions on exterior phenotype information from the Utah cohort generated with ClinPhen, CLiX, cTAKES, MedLEE, and MetaMapLite. MPSE’s precision and diagnostic yield amongst top-scoring probands is plotted in panel A of Fig. 1. Aside from MetaMapLite, all of the CNLP instruments’ outputs work nicely when used as inputs for MPSE, a truth made clear by the excessive restoration charges of sequenced and identified sufferers in comparison with selecting sufferers randomly for WGS. If MPSE was used to mechanically choose a quantity of NICU sufferers for sequencing equivalent in dimension to the Utah NeoSeq research (n = 65) from among the many 1838 whole sufferers screened, CLiX and ClinPhen would keep the NeoSeq research’s physician-mediated diagnostic yield (40%) all through the highest 50% of MPSE scores. This discovering accords nicely with our earlier publication, which confirmed excessive projected diagnostic yields from MPSE prioritization12.
Panels A, B show MPSE precision charges of sufferers manually chosen for WGS (Panel A) and diagnostic yield for the subset of circumstances identified by WGS (Panel B) utilizing completely different CNLP instruments. A CLiX-trained MPSE mannequin from the RCHSD cohort was utilized to phenotype information from 1838 College of Utah NICU sufferers generated by 5 completely different CNLP instruments. Panels C, D show precision and diagnostic yield utilizing MPSE fashions educated on 4 different information sorts (prognosis codes, lab checks, drugs, and all orders), in comparison with the corresponding HPO-based (CLiX) mannequin educated on the identical Utah cohort. A strong black reference line in every panel represents the precision or diagnostic yield anticipated from a mannequin that chooses candidates at random, whereas the black dashed line within the diagnostic yield graphs (Panels B, D) signifies the NeoSeq research’s 40% whole diagnostic yield. Determine generated with R ggplot2 software program.
Along with assessing yield amongst top-scoring probands, we additionally calculated cohort-wide efficiency metrics (see Supplementary Desk 3). After the “native” CLiX information, ClinPhen yields the next-best general predictions, with a mean 20.7-point distinction in MPSE rating between circumstances and controls (p = 2.2e−14), the best space beneath the ROC curve (AUROC = 0.91), and the best space beneath the PRC curve (AUPRC = 0.45). MetaMapLite exhibited the bottom efficiency, seemingly brought on by the relative dissimilarity between CLiX and MetaMapLite as seen within the low semantic similarity coefficients of CLiX-MetaMapLite time period units (Supplementary Fig. 1).
MPSE efficiency utilizing different information sorts
To find out whether or not non-phenotype information sorts might be used with MPSE, we examined MPSE fashions constructed utilizing prognosis codes, lab checks, and drugs and in contrast their predictive capability to phenotype-based fashions. A brief description of those information sorts is given in Supplementary Desk 4, and abstract statistics of commentary counts for every different information sort are proven in Supplementary Desk 5. MPSE fashions educated with different information sorts recovered fewer sequenced circumstances and diagnostic circumstances (Fig. 1B) amongst top-scoring probands than a corresponding phenotype-based mannequin however nonetheless carried out a lot better than a random mannequin. Among the many different information sorts, the ICD-10 primarily based mannequin yielded the most effective general predictions, approaching the efficiency of the CLiX-based mannequin (Supplementary Desk 3). Our evaluation means that non-phenotype structured information from affected person EHRs is much less efficient than CNLP-derived phenotype information at figuring out NICU sequencing candidates, however is a helpful and legitimate substitute for CNLP phenotype descriptions if these are usually not obtainable. Nevertheless, it must be famous that this evaluation was restricted to solely utilizing the presence/absence of a lab check, medicine order, and so on. (see Strategies) and never the check end result or order specs. An evaluation utilizing extra exact laboratory and medicine information might reveal larger MPSE efficiency when utilizing these different information sorts.
Diagnostic efficiency utilizing completely different CNLP instruments
Lastly, we assayed the final word utility of the CNLP-derived phenotype time period units generated by every device for medical molecular diagnostic actions. For these analyses, we used an Synthetic Intelligence (AI)-based gene prioritization device referred to as GEM11. Licensed from Material Genomics, by each RCHSD and the College of Utah, GEM is a business device that mixes HPO-based phenotype descriptions with WES and WGS sequences for speedy, AI-based diagnostic determination help. GEM was utilized by each RCHSD and the College of Utah for the unique prognosis of each sequenced proband within the datasets analyzed right here.
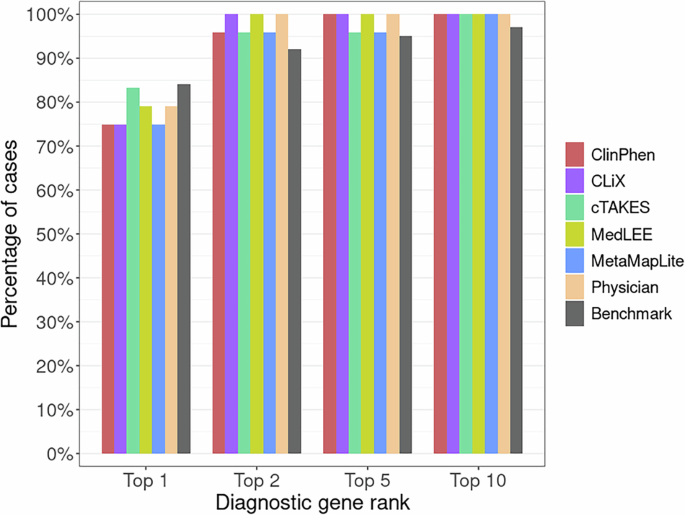
Comparability of GEM’s beforehand printed diagnostic efficiency to the possible Utah information reported right here offers a novel alternative each to reexamine GEM’s efficiency utilizing new, orthogonal information, and to assay the affect of utilizing completely different CNLP instruments on GEM’s diagnostic efficiency. These information are proven in Fig. 2. For reference, the unique GEM benchmark outcomes utilizing manually curated HPO time period units for 119 RCHSD probands11 have been added for ease of comparability. Determine 2 reveals the share of identified Utah NeoSeq23 probands the place the medical molecular diagnostic genotype was reported by GEM amongst its high 1st, 2nd, fifth, and tenth gene candidates.
Bars present the proportion of identified NeoSeq probands the place the true causal genes had been recognized by GEM among the many high 1st, 2nd, fifth, and tenth gene candidates. Every GEM run differed by the enter HPO time period lists, which had been made by extracting phenotypes from affected person medical notes utilizing CNLP (ClinPhen, CLiX, cTAKES, MedLEE, MetaMapLite) or handbook doctor assessment. The RCHSD benchmark (n = 119 sufferers) outcomes from the unique GEM paper are included for reference (redrawn with authors’ permission). Determine generated with R ggplot2 software program.
Two main conclusions emerge from these information. First, diagnostic efficiency utilizing CNLP-derived HPO information, whatever the device used to generate them, is almost equivalent to the efficiency obtained utilizing handbook time period units, and second, the outcomes proven listed here are extremely concordant with the unique printed benchmarking outcomes11. Furthermore, the College of Utah information offers a wholly potential orthogonal dataset, demonstrating transportability. These outcomes exhibit a strong synergy between the MPSE strategy and GEM. Specifically, that the identical HPO time period units utilized by MPSE for WGS prioritization could be instantly consumed by GEM for downstream diagnoses. Furthermore, Fig. 2 makes it clear that GEM can use HPO time period units, manually curated or CNLP derived, no matter device, with out compromising downstream diagnostic accuracy.
MPSE can determine sufferers who would profit from WGS inside the first 48 h of NICU admission
Our preliminary work and that introduced right here has demonstrated MPSE’s capability to precisely determine sequencing candidates by aggregating data from the whole lot of the affected person’s NICU keep. These findings underscore the device’s effectiveness in a retrospective context, utilizing all notes as much as the date at which the attending physicians place the order for WGS. Nevertheless, the final word check of worth lies in validating the real-time utility of MPSE. Early identification of sufferers who would profit from WGS, as quickly as attainable after NICU admission, might considerably improve care, enabling earlier illness prognosis and extra well timed interventions2,14,15.
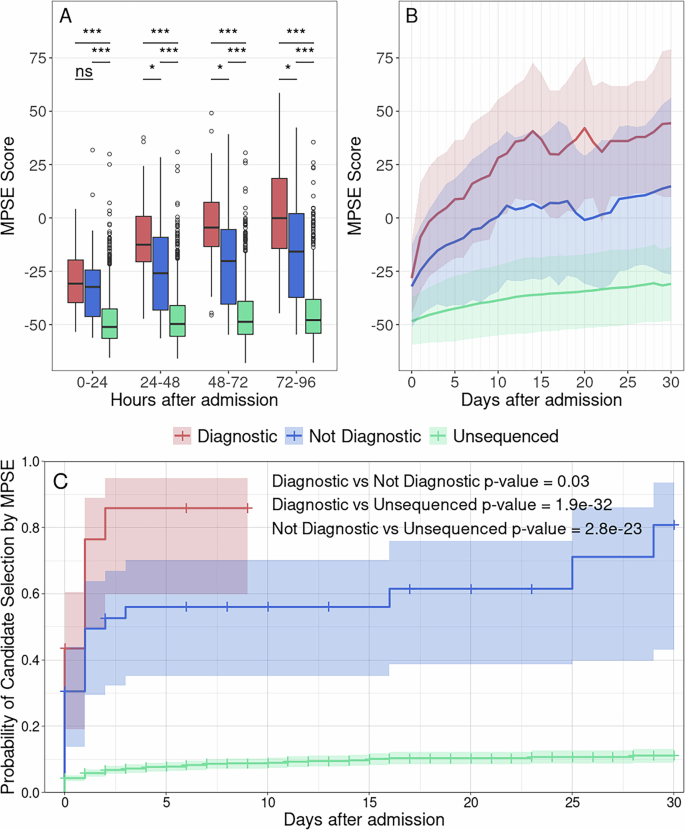
To measure MPSE’s real-time utility, we calculated every day MPSE scores for every affected person in our Utah cohort utilizing solely HPO phrases extracted with CLiX from medical notes current within the EHR at 24-h intervals, starting for the time being of their admission. Thus, every affected person had a collection of MPSE scores for every day spent within the NICU from admission to discharge. Longitudinal MPSE scores for sufferers who acquired a molecular prognosis by WGS (diagnostic), these for whom WGS didn’t determine a molecular prognosis (non-diagnostic), and sufferers who weren’t chosen for WGS (unsequenced) are summarized in Supplementary Desk 6 and plotted in Fig. 3 to assist visualize the change in MPSE rating over time amongst these teams. By the top of the primary day (0–24 h) within the NICU, each diagnostic and non-diagnostic sequenced circumstances had statistically considerably larger MPSE scores than did those that weren’t chosen for sequencing (unsequenced imply: −48.4; diagnostic imply: −32.1, p = 1.4e−5; non-diagnostic imply: −28.2, p = 9.3e−6). Moreover, diagnostic circumstances had considerably larger common MPSE scores than non-diagnostic sequenced circumstances starting 48 h post-admission (non-diagnostic imply: −24.7; diagnostic imply: −9.0; p = 0.018) and persevering with thereafter.
Panel A reveals MPSE rating distributions throughout the primary 96 h within the NICU for diagnostic (purple) and sequenced however non-diagnostic (blue) sufferers in addition to unsequenced NICU sufferers (inexperienced). Diagnostic and non-diagnostic sequenced sufferers had considerably larger MPSE scores than unsequenced sufferers starting 0–24 h after admission. Diagnostic sufferers had considerably larger MPSE scores than non-diagnostic sufferers starting at 48 h post-admission. Boxplot comparability significance ranges: *** (p p B reveals MPSE rating trajectories for these teams throughout the primary 30 days within the NICU. Strong traces present the imply MPSE rating per group and the shaded areas cowl one commonplace deviation from every imply. Panel C reveals the chance of at-risk sufferers being categorized as a WGS candidate by MPSE (i.e., MPSE rating >2 commonplace deviations above the unsequenced imply rating) as a operate of time. Cox proportional hazards regression evaluation confirmed the considerably elevated charge of MPSE candidate choice for diagnostic and non-diagnostic sufferers chosen for WGS in comparison with unsequenced sufferers, with hazards ratios of 18.8 (95% CI 11.6–30.6; p = 1.9e−32) and 9.8 (95% CI 6.2–15.3; p = 2.8e−23) respectively. Diagnostic sufferers had been additionally chosen by MPSE at the next charge than sequenced however non-diagnostic sufferers (HR = 2.0; 95% CI 1.1–3.9; p = 0.03). Determine generated with R ggplot2 software program.
Along with variations in MPSE scores between unsequenced, non-diagnostic, and diagnostic sufferers, there are additionally vital variations within the every day change in MPSE rating (day-N delta) between these teams. Each diagnostic and non-diagnostic sequenced circumstances noticed better common day-N delta than unsequenced controls all through the primary 30 days post-admission (Supplementary Desk 6). Importantly, the best distinction in MPSE rating enhance was noticed for the day-one delta, i.e. the change in MPSE rating through the first 24 h post-admission, with common diagnostic MPSE rating rising by 18.4 factors, common non-diagnostic MPSE rating rising by 7.7 factors, and common unsequenced MPSE rating rising by solely 2.7 factors. These day-one delta variations had been statistically vital for every subgroup comparability (diagnostic vs unsequenced p-value = 0.0015; non-diagnostic vs unsequenced p-value = 0.004; diagnostic vs non-diagnostic p-value = 0.027).
To realize additional perception into the temporal dynamics of MPSE’s predictive capabilities throughout the NICU keep, we estimated the proportional danger of being recognized as a WGS candidate by MPSE utilizing Cox proportional hazards regression evaluation24. The estimated chance and 95% CI of candidate choice by MPSE for diagnostic, non-diagnostic, and unsequenced sufferers is plotted in Panel C of Fig. 3. Utilizing a rule-of-thumb MPSE rating threshold (calculated individually for every day) of two commonplace deviations above the imply rating of unsequenced management sufferers, diagnostic sufferers had been flagged by MPSE at a considerably larger frequency and pace than each unsequenced sufferers (HR = 18.8, p = 1.9e−32) and non-diagnostic sequenced sufferers (HR = 2.0, p = 0.03). At 48 h post-admission, MPSE had already flagged 74% (17 of 23) of diagnostic sufferers, 47% (17 of 36) of non-diagnostic sequenced sufferers, and solely 6% (100 of 1773) of unsequenced sufferers. Moreover, by 9 days post-admission all diagnostic sufferers had been flagged by MPSE or had been censored on account of loss of life or NICU discharge, highlighting the pace at which MPSE was in a position to decide right medical motion for this group of sufferers. Supplementary Desk 7 incorporates the every day rating threshold, the variety of candidates assessed by MPSE, and the cumulative variety of sufferers who reached the rating threshold as a part of this longitudinal evaluation.