Info on ACA/ECA gene members of the family and phylogenetic relationships in lettuce
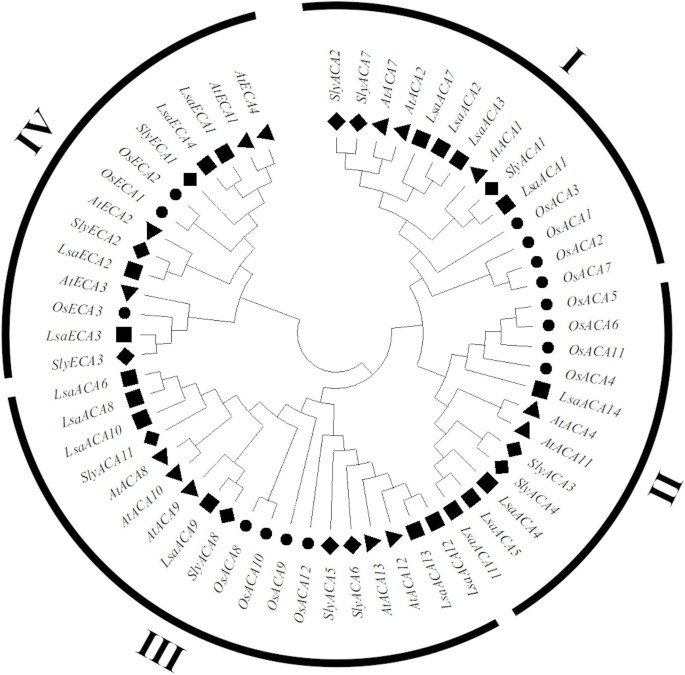
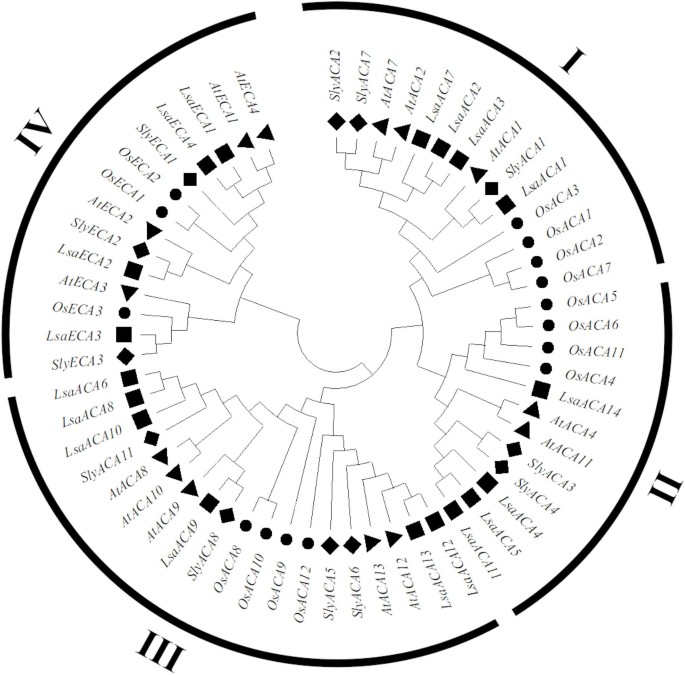
This examine recognized 18 members of the lettuce ACA/ECA household utilizing bioinformatics strategies. The ACA/ECA protein household could be categorized into 4 extremely conserved evolutionary branches (I-IV), amongst which department III incorporates probably the most prevalent kinds of ACA/ECA in lettuce, rice, tomato, and Arabidopsis (Fig. 1).
The molecular weights of lettuce ACA/ECA members of the family vary from 123993.02 Da (LsaACA5) to 104733.5 Da (LsaACA3), with a median molecular weight of 114033.71 Da and a median variety of amino acids of 1039.83. The theoretical isoelectric factors vary from 5.16 (LsaECA2) to 7.77 (LsaACA14), with a median worth of 6.13. This consists of 15 acidic proteins (pI 7). The general common hydrophilicity ranges from − 0.017 (LsaECA4) to 0.248 (LsaECA3), with a median of 0.107. The instability coefficient ranges from 27.42 (LsaACA7) to 40.03 (LsaACA9), with a median of 34.50 (Desk 1).
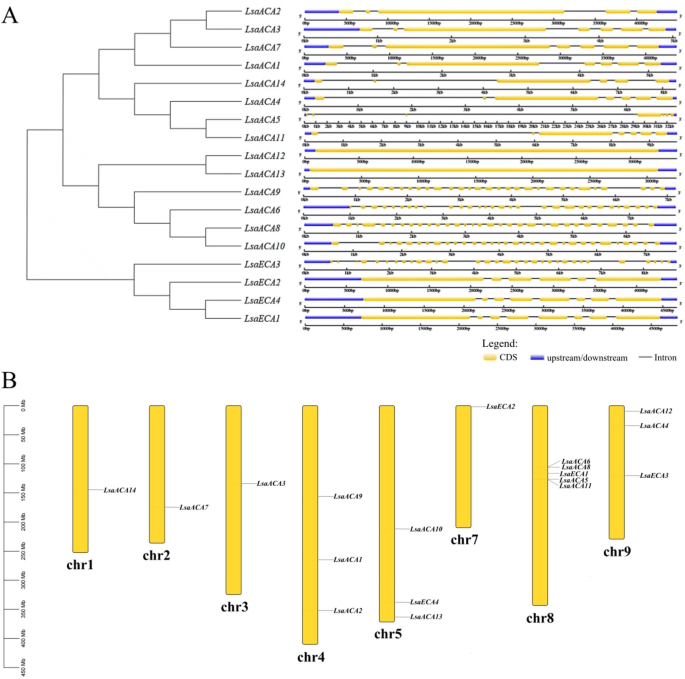
Lettuce ACA/ECA gene construction and gene localization on chromosomes
The outcomes confirmed noteworthy variations within the variety of exons among the many members of the lettuce ACA/ECA gene household. As an example, LsaACA13 and LsaACA12 include solely a single exon every, whereas LsaECA3, LsaACA10, LsaACA8, LsaACA6, and LsaACA9 include greater than 30 exons every, and different lettuce ACA/ECA genes have 5–10 exons. There are minimal variations within the quantity and place of exons/introns amongst members inside the similar department (Fig. 2).
This examine discovered that 18 ACA/ECA genes are erratically distributed on the eight chromosomes of lettuce, with chromosome 8 containing the best variety of ACA/ECA household genes (5), adopted by chromosomes 4, 5, and 9, every containing three ACA/ECA genes. Moreover, the positions of LsaACA8 and LsaACA6, in addition to LsaACA5 and LsaACA11 on chromosome 8, are comparatively shut (Fig. 2), indicating that they might be homologous genes ensuing from fragment replication.
The gene construction and chromosome localization of the ACA/ECA gene household in lettuce. (A) Exon-intron constructions of LsaACA/ECA genes. (B) LsaACA/ECA genes on lettuce chromosomes. The yellow rectangular bars characterize the lettuce chromosomes, and the 0-450 Mb scale represents the chromosome size. The gene ID of LsaACA/ECA is highlighted in black for the corresponding chromosome.
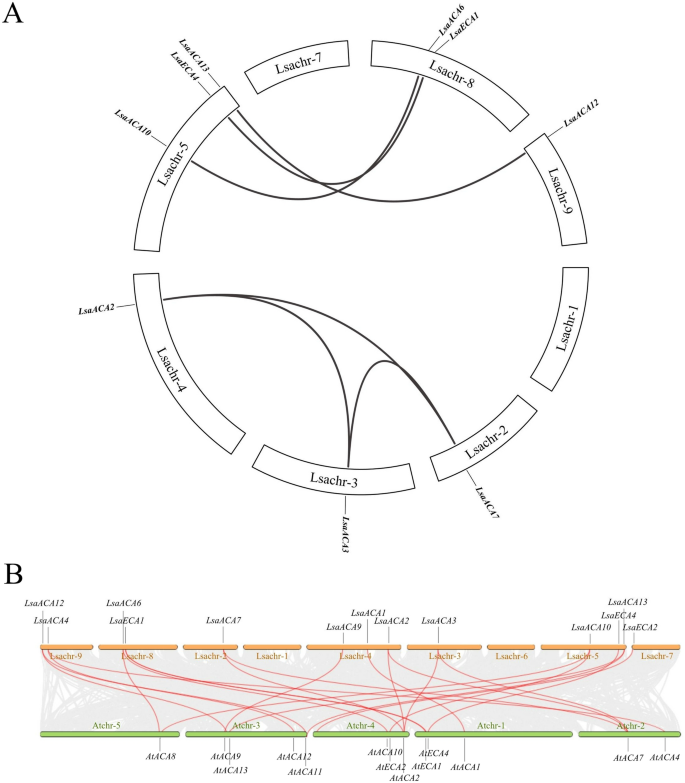
Collinearity evaluation of ACA/ECA gene members of the family in lettuce
Throughout the lettuce ACA/ECA gene household, six pairs of genes exhibited collinearity, particularly, LsaACA7 with LsaACA3 and LsaACA2, LsaACA3 with LsaACA2, LsaACA10 with LsaACA6, LsaECA4 with LsaECA1, and LsaACA13 with LsaACA12 (Fig. 3). This means that the lettuce ACA/ECA gene has undergone segmental duplication between genes all through its evolution.
There are 21 pairs of ACA/ECA genes exhibiting collinearity between lettuce and Arabidopsis (Fig. 3), indicating a sure degree of homology inside the ACA/ECA gene household in each species.
Collinearity evaluation of ACA/ECA genes in lettuce and in Arabidopsis. (A) Collinearity evaluation of ACA/ECA genes in lettuce. The black traces characterize segmental duplication occasions between genes. (B) Collinearity evaluation of ACA/ECA genes between lettuce and Arabidopsis. The chromosomes are distinctly coloured. The grey traces within the background point out the collinear blocks inside lettuce and Arabidopsis genomes. The purple traces join syntenic ACA/ECA gene pairs.
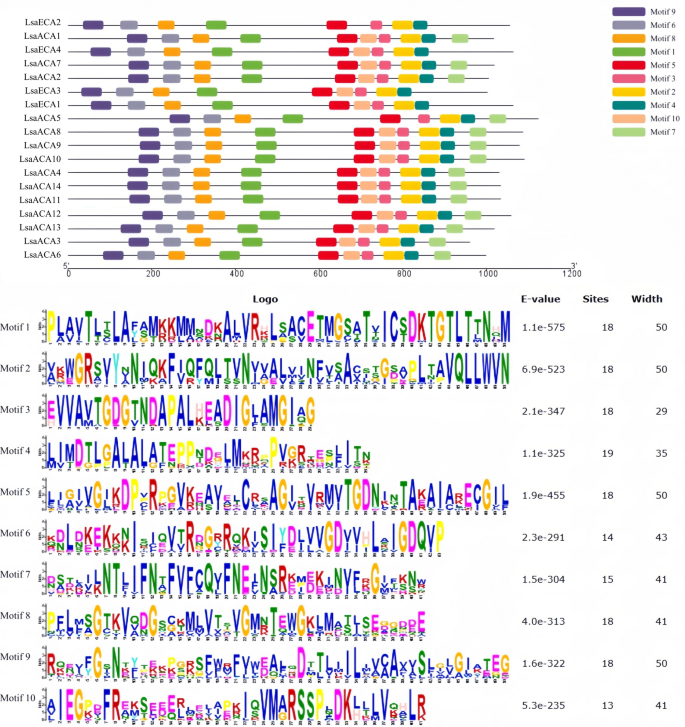
Evaluation of protein-conserved domains of lettuce ACA/ECA gene household
Utilizing the net instruments Pfam and MEME, we analyzed the conserved domains of lettuce ACA/ECA household proteins and obtained 10 conserved motifs, particularly, Motif 1-Motif 10. Among the many 18 members of the lettuce ACA/ECA household proteins, LsaECA4, LsaECA3, and LsaECA1 lack Motif 7, LsaACA5 lacks Motif 10, and LsaECA2 lacks Motif 7 and Motif 10 (Fig. 4). It’s speculated that ACA/ECA practical differentiation throughout evolution might have led to variations within the conserved motif composition of lettuce ACA/ECA members of the family. Additional evaluation revealed that Motif 1 incorporates a repeatedly extremely conserved D-Ok-T-G-T-L-T sequence at its finish, Motif 2 incorporates a repeatedly extremely conserved L-L-W-V-N sequence at its finish, and Motif 8 incorporates 4 discontinuous extremely conserved glycine (G) residues (Fig. 5).
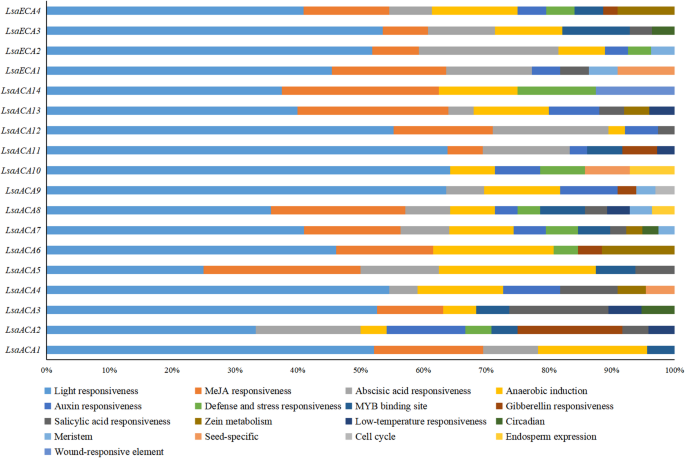
Evaluation of Cis-acting parts of lettuce ACA/ECA gene household
It was discovered that the lettuce ACA/ECA gene household incorporates a big variety of photoresponsive parts, with 16 ACA/ECA genes containing anaerobic induction parts, 14 ACA/ECA genes containing methyl jasmonate response parts and abscisic acid response parts, 12 ACA/ECA genes containing auxin response parts, and 10 ACA/ECA genes containing salicylic acid response parts (Fig. 6). Subsequently, the above outcomes point out that the lettuce ACA/ECA gene household can reply to numerous hormones or environmental indicators and may take part in associated regulation and physiological processes.
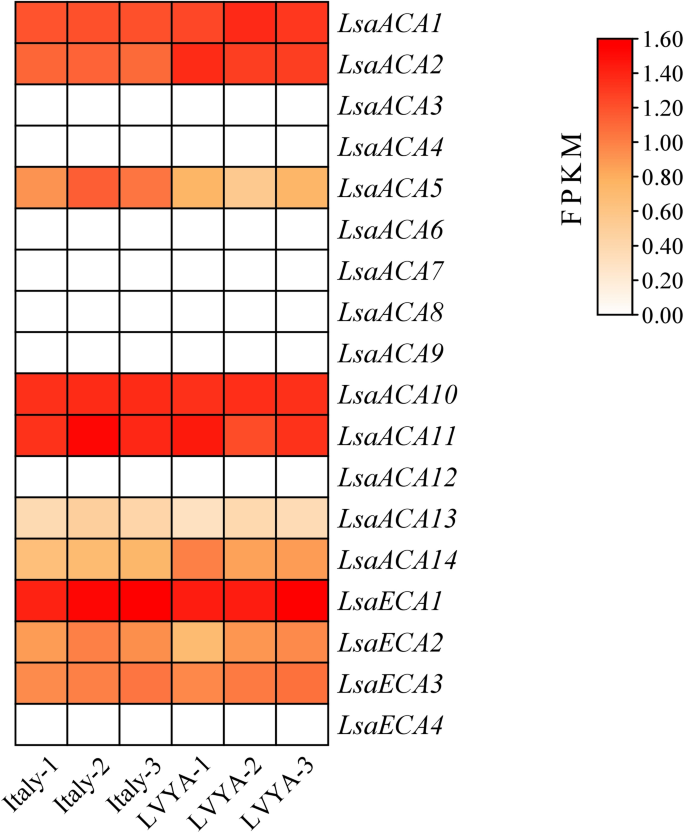
Expression of ACA/ECA gene household in lettuce leaves
Based mostly on the earlier transcriptome sequencing outcomes obtained by our analysis group, goal gene expression information had been collected and analyzed utilizing TBtools to create an digital expression heatmap. The outcomes confirmed that the expression ranges of the LsaACA1, LsaECA1, LsaACA2, LsaACA10, and LsaACA11 genes within the leaves of two lettuce varieties had been comparatively excessive (Fig. 7). In contrast with LsaECA1 and LsaACA1, LsaACA2 and LsaACA11 include stress response parts, and a larger variety of phytohormone responsive parts, LsaACA10 incorporates stress response parts too (Fig. 6). Subsequently, these three genes had been chosen on this examine and their capabilities had been validated utilizing transient silencing.
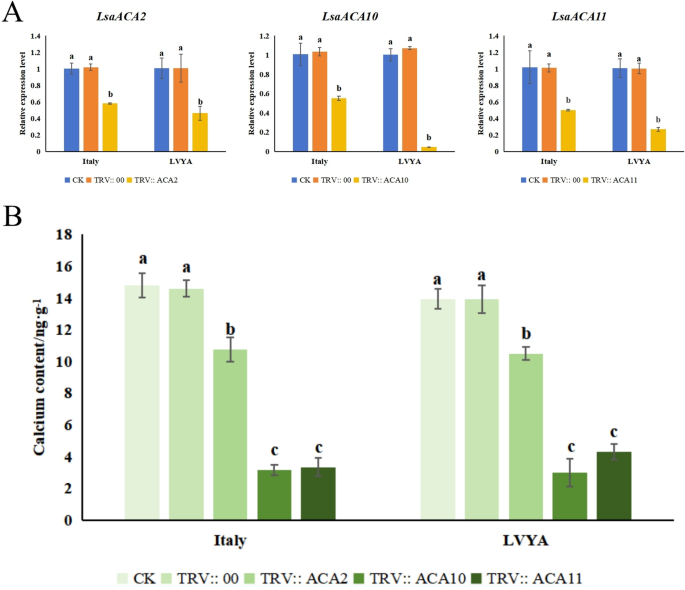
VIGS silence impact detection
On the twenty first day after an infection, new leaves contaminated with CK, empty TRV:: 00, TRV:: ACA2, TRV:: ACA10, and TRV:: ACA11 had been collected to establish the TRV virus and analyze the expression of ACA genes. In the meantime, the leaf phenotype of optimistic management (TRV:: PDS) was noticed. The outcomes indicated that the TRV virus was detected within the newly leaves (Supplementary Determine S1), the brand new leaves within the optimistic management take a look at confirmed albino signs (Supplementary Determine S2), and the expression ranges of ACA2, ACA10, and ACA11 within the leaves of two lettuce varieties had been considerably decrease than within the management. Amongst them, the expression ranges of ACA2, ACA10, and ACA11 in “Italy” lettuce decreased by 43.14%, 47.12%, and 50.50%, respectively, in comparison with the management (TRV:: 00). The expression ranges of ACA2, ACA10, and ACA11 in “LVYA” lettuce decreased by 54.46%, 96.26%, and 73.01%, respectively, in comparison with the management (TRV:: 00). Concurrently, the calcium content material within the leaves of each lettuce varieties was notably decreased compared to the management. The calcium content material within the leaves of “Italy” lettuce decreased by 26.36–78.38% in comparison with the management (TRV:: 00), whereas the calcium content material within the leaves of “LVYA” lettuce decreased by 24.60 -78.42% in comparison with the management (TRV:: 00) (Fig. 8 and supplementary Determine S3).
As well as, silencing the ACA gene in lettuce exacerbated the prevalence of tipburn in leaves (Fig. 9). The variety of tipburn vegetation, the utmost variety of tipburn leaves per plant, and the typical tipburn price of each types of lettuce had been larger than within the management. Amongst them, the lettuce contaminated with TRV:: ACA10 had probably the most extreme tipburn, with all six contaminated vegetation exhibiting tipburn, and the typical tipburn price of the tipburn vegetation was larger than that of the management and different experimental teams. The variety of tipburn vegetation in “Italy” lettuce elevated by 66.67–100.00% in comparison with the management (TRV:: 00), and the utmost variety of tipburn leaves per plant elevated by 120.00 -140.00% in comparison with the management (TRV:: 00). The typical tipburn price of the tipburn vegetation elevated by 118.65–131.80% in comparison with the management (TRV:: 00). The variety of tipburn vegetation and the utmost variety of tipburn leaves per plant in “LVYA” lettuce elevated by 150.00–200.00% in comparison with the management (TRV:: 00), and the typical tipburn price of diseased vegetation elevated by 180.61–243.88% in comparison with the management (TRV:: 00) (Desk 2).