Protein design and expression
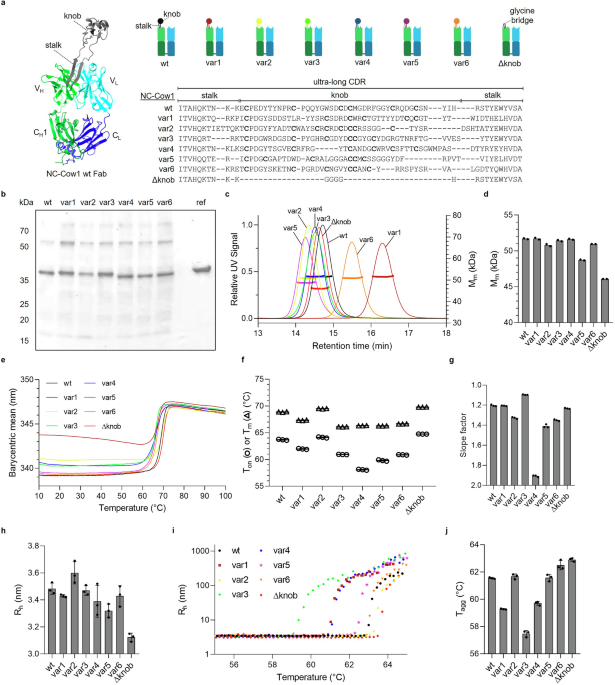
The Fab fragments with ulCDRs had been obtained through the use of the VH and VL domains of NC-Cow1 (PDB:6OO0) fused to bovine CH1 and CL from BOV-7 (PDB:6E9U), as described earlier than9. The design of the ulCDR variants is described within the outcomes part. Plasmids had been obtained by industrial gene synthesis from GeneArt (Thermo Fisher)9. The identical method was used for acquiring the plasmids for recombinant human EGFR and Nkp30 (extracellular domains with C-terminal 6-His tag) and of the dad or mum Fab fragments of var113, var3, and var424. The plasmids for the HIV-1 Env antigen (BG505 SOSIP.664 gp140-his) and furin had been kindly supplied by John P. Moore at Cornell College9. Bigger quantities of the plasmids had been obtained by purification with Midiprep and Maxiprep kits (Thermo Fisher) from in a single day cultures of reworked XL1-Blue cells, adopted by sequencing to substantiate the proper inserts. All Fab fragments had been produced by transient transfection of Expi293™ cells (Thermo Fisher) grown in Expi293™ expression medium at 37 °C with 8% CO2. The cells had been transfected with 0.5 µg plasmid per 1 mL of cell suspension (2:1 LC/Fd plasmid ratio) utilizing the FectoPRO® DNA transfection equipment (Polyplus) following the producer’s protocols. For producing the HIV-1 Env antigen, cells had been transfected with 1 µg plasmid per 1 mL of cell suspension (4:1 BG505 SOSIP.664/furin plasmid ratio) utilizing the ExpiFectamine™ 293 transfection equipment in accordance with the producer’s protocols. Recombinant human EGFR and Nkp30 had been produced by transient transfection of ExpiCHO™ cells (Thermo Fisher) grown in CHOgro® Expression Medium (Mirus) at 37 °C with 8% CO2. The cells had been transfected with 1 µg plasmid per 1 mL of cell suspension utilizing the CHOgro® Excessive Yield Expression System (Mirus) following producers’ pointers. Instantly after the transfection of ExpiCHO™ cells, the temperature was switched to 32 °C till harvest. Cell supernatants from all transfections had been collected by centrifugation 4–8 days after transfection. For purification of proteins, an ÄKTA pure (Cytiva) was used at 4–5 °C. The secreted Fab fragments had been purified from the cell supernatants by affinity chromatography utilizing a self-packed column with CaptureSelect™ LC-lambda (ung) affinity matrix (Thermo Fisher). After a washing step with phosphate-buffer saline (PBS), the sure pattern was eluted with 0.1 M glycine pH 3.0 into 1/5 quantity of 1 M tris pH 8.5. His-tagged proteins had been purified with a HisTrap Excel column (Cytiva). After a washing step with washing buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4), the sure samples had been eluted with elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). After affinity chromatography, the samples had been additional purified by size-exclusion chromatography with a Superose 6 Improve 10/300 GL column (Cytiva) within the case of HIV-1 Env antigen, and a HiPrep Sephacryl S-200 HR column (Cytiva) within the case of all different proteins, utilizing PBS as a operating buffer. Lastly, the purified proteins had been concentrated with Centricon® centrifugal filter gadgets (Millipore), and the samples had been frozen at −80 °C for later use. All samples had been saved in PBS pH 7.4.
Gel electrophoresis
Cell supernatants and purified proteins had been analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis utilizing SERVAGel™ TG PRiME™ 4–20% gels and a Twin Coloration Protein Commonplace III (SERVA Electrophoresis GmbH). Samples had been combined with 4× Laemmli Pattern Buffer (Bio-Rad), and for the decreasing gel, DTT (Merck) was added to a focus of 200 mM. Samples had been incubated at 95 °C for five min earlier than being transferred into the gel.
Dimension-exclusion chromatography coupled to multi-angle gentle scattering (SEC-MALS)
A Waters 2695 Separation Module HPLC related to a Waters 2487 Twin Absorbance UV Detector (Waters), a miniDAWN TREOS MALS detector (Wyatt Know-how), and an Optilab rEX refractive index detector (Wyatt Know-how) was used for many SEC-MALS measurements. For the measurements of var1 or var1PA combined with HIV Env trimer, var3, var3PA, var4 or var4PA combined with EGFR, and of var1PA, var3PA, and var4PA alone, an Arc HPLC Quaternary Solvent Supervisor-R (Waters), an Arc HPLC Pattern Supervisor FTN-R (Waters), a 2489 UV/Vis Detector (Waters), a Fraction Supervisor (Waters), a miniDAWN TREOS MALS detector and an Optilab refractive index detector (Wyatt Know-how) had been used. As a operating buffer, PBS with 200 ppm sodium azide was used, and the circulate fee was 1 mL/min. A Superdex 200 Improve 10/300 GL column (Cytiva) was used for separation. The chromatograms had been collected and evaluated utilizing the Astra software program v8.1.2 (Wyatt Know-how). For molar mass dedication, the UV sign with theoretical extinction coefficients was used as a focus supply within the case of the samples containing solely Fab fragment, and the RI sign with a relentless dn/dc worth was used as a focus supply for all different samples.
Dynamic gentle scattering (DLS)
A DynaPro plate reader (Wyatt Know-how) was used for dynamic gentle scattering measurements. The measurements had been carried out in 384 spherical nicely low-volume microplates (Aurora Microplates) in triplicates utilizing 30 µl of pattern which was sealed with a couple of µl of silicone oil. Previous to measurement, the plates had been centrifuged for two min at 2000 rpm. For knowledge assortment and processing, the DYNAMICS software program model 8.2 (Wyatt Know-how) was used. The obvious hydrodynamic radius (Rh) and the onset temperature of aggregation (Tagg) had been decided utilizing a protein focus of 0.5 mg/mL. For isothermal measurements, 10 acquisitions with an acquisition time of 5 s had been used for every measurement. For figuring out Tagg, a temperature ramp of 0.1 °C/min was utilized from 25 to 70 °C, and one measurement included 3 acquisitions of three s. Tagg was calculated from the rise in Rh throughout heating by the DYNAMICS software program.
Differential scanning fluorimetry (DSF)
Thermal protein unfolding was assessed utilizing a SUPR-DSF (Protein Steady) system that measures intrinsic protein fluorescence depth. The measurements had been carried out in 384-well thin-wall Arduous-Shell PCR plates (Bio-Rad) in triplicates utilizing 10 µl of pattern sealed with Microseal ‘B’ PCR Plate Sealing Movie (Bio-Rad). A protein focus of 0.5 mg/mL was used for measurements. A temperature ramp of 1 °C/min was utilized from 10 to 105 °C. Samples had been excited at 280 nm, and the barycentric imply inside a variety of 310 nm to 390 nm was plotted in opposition to the temperature. The obvious onset temperature of unfolding Ton and the obvious melting temperature Tm had been calculated with the SUPR Suite software program v3.0 (Protein Steady). For evaluating the slopes of the melting curves, a sigmoidal Boltzmann match was carried out utilizing GraphPad Prism model 8.0.1 for Home windows, GraphPad Software program, Boston, Massachusetts USA, www.graphpad.com.
Hydrophobic interplay chromatography (HIC)
A Dionex Summit 2 system (Dionex) related to a UVD170U detector (Dionex) was used. All samples had been measured as duplicates utilizing a Proteomix® HIC Butyl-NP5, 5 μm column (Sepax Applied sciences). Two operating buffers had been used: Buffer A contained 1.8 M ammonium sulfate and 0.1 M sodium phosphate, pH 5, and Buffer B contained 0.1 M sodium phosphate, pH 5. Samples had been ready by mixing proteins in PBS with Buffer A to lead to an ammonium sulfate focus of 1 M. Every run was achieved with a circulate fee of 0.5 mL/min, consisting of a column equilibration step with 100% Buffer A for 25 min previous to pattern injection, a gradient from 0% Buffer B to 100% Buffer B inside 40 min, and one other 10 min with 100% Buffer B.
Biolayer interferometry (BLI)
The binding affinity of Fab fragments to antigens was measured utilizing a BLItz system (FortéBio). After an preliminary baseline of 30 s in PBS, antigen containing a His-tag was captured on an Octet® Anti-Penta-HIS (HIS1K) biosensor (Sartorius) for 300 s at a focus of 25 ng/µL. Then, after a second baseline of the 30 s, the biosensor pattern with immobilized antigen was positioned into an answer containing Fab fragment, and the affiliation was recorded for 180 s. After the affiliation step, dissociation was measured for 300 s by putting the sensor into an answer of PBS. Every Fab fragment was measured for antigen binding at 4 totally different concentrations and normalized to a pattern containing solely PBS. OkD values had been calculated with becoming capabilities utilizing the BLItz Professional software program (FortéBio).
Round dichroism (CD) spectroscopy
Far- and near-UV CD spectra of all protein constructs had been acquired on a J-1500 CD spectrometer (JASCO) outfitted with a temperature management system coupled with multi-position cells. Close to-UV CD samples had been acquired at a focus of 1 mg/ml utilizing a quartz cell with a 1 cm path size. The wavelength was assorted from 250 to 350 nm with a 0.1 nm step and an acquisition time of three s per level. For every CD spectrum, three scans had been averaged and smoothed utilizing the Savitzky–Golay technique. Far-UV CD spectra had been obtained on 0.02 mg/ml samples utilizing a quartz cell with a 0.1 cm path size. The wavelength was assorted from 200 to 280 nm with 0.1 nm step and acquisition time of three s per level. For every CD spectrum, three scans had been averaged and smoothed utilizing the Savitzky–Golay technique38. All spectra had been processed and analyzed utilizing Spectra Evaluation software program (JASCO). Secondary construction evaluation was carried out with Bestsel software program24 with a area between 200 and 250 nm included within the match39,40,41,42.
Microfluidic modulation spectroscopy (MMS)
Mid-infrared spectra had been recorded with an Aurora (RedShiftBio) utilizing a protein focus of 1.4–1.6 mg/mL in PBS. Earlier than measurement, the totally different ulCDR variants had been dialyzed in opposition to Dulbecco’s phosphate buffered saline (VWR) in a Pierce 96-well Microdialysis plate (ThermoFisher) by following the producer’s protocol. This was to make sure an optimum buffer alignment between the pattern and reference buffer. The measurements had been carried out in 96-well spherical backside plates (Corning) sealed with Zone-Free™ Sealing Movies (Excel Scientific). Every pattern was measured in three replicates, and normalized common absolute absorbance spectra and second by-product spectra had been calculated utilizing delta software program (RedShiftBio). For absolutely the absorbance spectra, a nominal match displacement issue of 0.6 and a match vary of 1720 cm−1 to 1680 cm−1 had been used. For the second by-product spectra, Savitzky-Golay smoothing was utilized utilizing a window of 19 wavenumbers38.
Nuclear magnetic resonance (NMR) spectroscopy
Samples for NMR had been ready by the addition of 5% v/v 2H2O to 500 μL of a 1 mg/ml answer of proteins and transferred to five mm precision NMR tubes (Wilmad). All NMR spectra had been acquired at 25 °C on a 600 MHz Avance NEO spectrometer (Bruker) outfitted with a 5 mm triple resonance TCI cryoprobe and a temperature management unit. For every pattern, 1D 1H spectra had been acquired utilizing a typical zggpwg Bruker pulse sequence. The spectra had been acquired and processed utilizing Bruker Topspin 4.0.8 (Bruker). Pearson correlation was used to calculate the similarity index between proteins, the place areas of the spectra that comprise non-protein elements had been discarded from evaluation.
Hydrogen–deuterium trade coupled to mass spectrometry (HDX-MS)
Labeling and measurements had been carried out utilizing an HDX setup from Waters. This features a PAL RTC Autosampler (LEAP Applied sciences), a UHPLC with μBinary Pump and Auxiliary Pump (Waters), the HDX Supervisor of separate column ovens for the pepsin column and analytical column (Waters), and a Synapt XS (Waters). For again trade, a myoglobin answer of 20 µM in water and a myoglobin answer of 20 µM in D2O had been ready. For full deuteration, the myoglobin in D2O was shaken at 35 °C for 1.5 h, and 170 mg NaCl was added to scale back the freezing level. Protein options had been saved within the quench tray at 1 °C below nitrogen. Three microlitres of protein answer had been injected into the labeling vial. Then, 57 μL of labeling buffer (pD 7.4, 5 mM Ok2HPO4, 5 mM KH2PO4, 150 mM KCl in D2O) or equilibration buffer (similar as labeling buffer, however with H2O as a substitute of D2O) had been added and allowed to react for the set time at 20 °C. Fifty microlitres of the response answer had been transferred to the quench vial within the quench tray containing 50 μL of quench buffer (pH 2.3, 50 mM Ok2HPO4, 50 mM KH2PO4, 1 M TCEP-HCl, 0.7 M NaOH, 4 M guanidine-HCl in H2O) at 1 °C. Fifty microlitres of quenched pattern had been injected right into a BEH pepsin column (Waters) earlier than coming into an ACQUITY UPLC BEH C18 VanGuard Precolumn (Waters) coupled to an ACQUITY UPLC BEH C18 column (Waters). Fifty microlitres of the quenched pattern had been injected right into a BEH pepsin column, 5 μm, 30 × 2.1 mm, 300 Å (Waters) and flowed for 1 min with a circulate of 75 μL/min, adopted by 3 min with a circulate of 200 µL/min with 0.2% formic acid in H2O at 20 °C earlier than coming into an ACQUITY UPLC BEH C18 VanGuard Precolumn, 1.7 μm, 5 × 2.1 mm, 130 Å (Waters) coupled to an ACQUITY UPLC BEH C18 column (Waters) at 1 °C. Thereafter, it began a gradient as proven in Desk 2 with the eluent A (H2O, adjusted with formic acid to pH 2.5) and eluent B (acetonitrile with 0.3% formic acid) over the entice column to the analytical column ACQUITY UPLC BEH C18, 1.7 μm, 150 × 1 mm, 130 Å (Waters) at 1 °C. The circulate fee was 45 μL/min over the whole time. The gradient is a developed and upscaled model of a beforehand printed technique43. A Waters ESI supply was used for ionization with the next settings: capillary voltage of three.0 kV, supply temperature of 90 °C, sampling cone of fifty.0 V, supply offset of 20.0 V, desolvation temperature of 250 °C, cone gasoline circulate of 100 L/h, desolvation gasoline circulate of 550 L/h, and nebulizer gasoline strain of 6 bar. The MS technique was UDMSe, with argon because the collision gasoline and nitrogen because the drift gasoline. The UDMSe technique (Desk 3) was developed by taking particular person collision energies for the cost states and mass-to-charge ratios primarily based on a earlier publication44. The wave velocity was ramped linearly from 1500 m/s to 450 m/s with a relentless wave peak of 40 V, a relentless helium gasoline circulate of 180 mL/min, and a relentless drift gasoline circulate of 90 mL/min. As LockSpray for recalibration, an answer of two ng/μL leucine enkephalin (Waters Company) in 50:50 acetonitrile:water with 0.1% formic acid was infused. The labeled protein samples had been measured 5 occasions for every time level at 0.25, 2.5, 13.75, 25, and 250 min, and the protein samples with equilibration buffer with time level 0 min had been measured with n = 2 for every protein conformation. All measurements with a time level of 0 min had been mixed as reference. For again trade, the myoglobin pattern in water was measured at n = 4 and t = 0 s, and the deuterated pattern was measured at n = 3 and t = 150 s. The analysis of the analytes was carried out utilizing ProteinLynx International Server and DynamX (Waters). The sequence used for the analysis was that of NC-Cow1 Fab Δknob for all variants. Primarily based on the peptide fragmentation sample, a rating threshold of 6.60 was chosen for the recognized peptides. For myoglobin (back-exchange), the rating threshold was 7.70. The depth threshold was 1000 for the peptides, with a mass error of a most of 10 ppm, 0.11 fragments per amino acid, a sum product depth of 470, and one performed product45. The chromatographic indicators between 2.6 min and 14.65 min had been evaluated. The cluster knowledge from DynamX was additional analyzed utilizing an Excel sheet developed in-house to generate tuptake plots and butterfly plots. This resulted in a sequence protection of 64.8% with 16 peptides for the Fd and a sequence protection of 83.3% with 16 peptides for the LC with a median again trade of 49%.
Molecular dynamics (MD)
The construction of a bovine Fab NC-Cow1 (PDB:6OO0) was used on this research. We carried out molecular dynamics simulations for 4 totally different constructs. Within the first system (wt), we used the variable segments of the NC-Cow1 (VH and VL). We changed the knob of the ulCDR with 4 glycines within the second system (Δknob). Within the third and fourth methods (var4, var5), we changed the 2 stalk areas instantly earlier than and after the knob of the ulCDR with the var4 and var5 variations, respectively. For the alternative, we used the Modeler program46. To check the reliability of the alternative modeling, we used two totally different preliminary fashions for var4. They each present related ends in the simulations.
All of the constructs had been positioned in a cubic field simulation utilizing the CHARMM-GUI internet server47. Sodium and chloride ions had been added to make a 150 mM ion focus. The all-atom CHARMM36m drive discipline was used to check the dynamics of protein, glycan, and ions with the TIP3P express mannequin for water molecules48. MD trajectories had been analyzed utilizing MDAnalysis and VMD49,50.
All simulations had been carried out utilizing GROMACS VERSION 2021.351. The preliminary setups had been minimized for 5000 steps with the steepest descent technique and later equilibrated for 500 ps in a canonical (NVT) ensemble and afterward for 7 ns in an isothermal-isobaric (NPT) ensemble below periodic boundary circumstances. The positional restraints on initially 4000 kJ mol−1 nm2 nonhydrogen protein atoms had been steadily launched throughout equilibration. Lengthy-range electrostatic interactions had been handled with the Particle-mesh Ewald summation52 with cubic interpolation and a 0.12-nm grid spacing. Throughout equilibration, the time step was first 1 fs and was then elevated to 2 fs throughout the NPT equilibration. The LINCS algorithm was used to repair all bond lengths53. In the course of the equilibration section, fixed temperature and strain had been established with a Berendsen thermostat, mixed with a coupling fixed of 1.0 ps and an isotropic Berendsen barostat, respectively54. The Berendsen thermostat and barostat had been changed by a Nosé–Hoover thermostat and a Parrinello-Rahman barostat throughout the manufacturing runs55,56. Evaluation was carried out on the manufacturing simulations. Three simulations had been carried out for every assemble (see Supplementary Desk 3 for the outline of the simulations).
Statistics and reproducibility
All experiments are reproducible. The imply values reported are derived from technical replicates. Statistical comparability was used just for the evaluation of the HDX-MS knowledge and was carried out utilizing a typical and out there software program, Deuteros 2.0, for significance testing of HDX-MS knowledge.
Reporting abstract
Additional info on analysis design is obtainable within the Nature Portfolio Reporting Abstract linked to this text.