Moral approval of animal and human research
The mouse and human tissue research described on this work complied with all related moral rules. The primary mouse fgTiO2 feeding research (i.e., Mouse Research 1) was accredited by the Grasslands Animal Ethics Committee (Palmerston North, New Zealand) in accordance with the New Zealand Animal Welfare Act 1999. The second mouse fgTiO2 feeding research (i.e., Mouse Research 2) was carried out at Cambridge beneath the authority of Dwelling Workplace private license quantity PIL IEDB62633, and venture license quantity PPL PF86EABB1. Following written knowledgeable consent, and after approval from the Regional Moral Evaluation Board, Linköping, Sweden, human surgical specimens containing Peyer’s patches have been collected from the neo-terminal ileum from sufferers with Crohn’s illness or colonic most cancers. Research utilizing human tissues at Cambridge have been additionally accredited by the UK NHS Well being Analysis Authority, North West—Higher Manchester East Analysis Ethics Committee, REC reference 18/NW/0690.
Mouse research 1
Meals-grade, anatase TiO2 particles (fgTiO2) have been bought from Sensient Colours (St. Louis, MO, USA). Physico-chemical characterisation of those particles confirmed a particle diameter distribution of ~ 50–300 nm, with a median diameter of ~ 130 nm1. As described and characterised beforehand15, to allow profitable oral supply, the usual rodent food regimen American Institute of Diet (AIN)−76A was supplemented with 0.000625%, 0.00625% or 0.0625% (w/w) fgTiO2 (translating to ~1, 10 or 100 mg fgTiO2/kg body-weight/day, respectively15). These diets have been ready by Analysis Diets (New Brunswick, NJ, USA), with straight AIN-76A with out fgTiO2 supplementation used because the unfavorable management food regimen. Forty-eight, six-week-old mice (Mus musculus C57BL/6 J, 50:50 male:feminine) have been sourced from the AgResearch Ruakura Small Animal Colony and randomly cut up into 4 feeding teams (0.00%, 0.000625%, 0.00625% or 0.0625% (w/w) fgTiO2/AIN-76A, n = 6 male and n = 6 feminine per group) with regular feed consumption and weight-gain confirmed bi-weekly15. Mice have been housed beneath a typical 12 h mild, 12 h darkish cycle, with a temperature maintained between 20 and 24 °C and a relative humidity of 40–60%. After eighteen weeks feeding, mice have been euthanised by CO2 asphyxiation and cervical dislocation. The gastrointestinal tract was shortly eliminated and positioned in chilly phosphate-buffered saline (PBS). Ileal tissues containing essentially the most distal Peyer’s patches (i.e., closest to the ileal/caecal junction) and adjoining caecal patches have been excised and frozen on a chilly, chrome steel bar embedded in dry ice. Tissues have been then transferred into cryomolds stuffed with pre-cooled optimum chopping temperature compound (OCT) and transferred to −80 °C for storage. In earlier work utilizing tissues from the identical animals, we established that fgTiO2 consumption and supply into Peyer’s patches was not considerably completely different between female and male animals15. For these causes and to maximise statistical energy, knowledge throughout men and women have been mixed and anlaysed collectively within the offered work. Throughout evaluation, intercourse data for particular person tissue sections was not retained so the offered knowledge can’t be disaggregated by intercourse.
Mouse research 2
Feminine mice (Mus musculus C57BL/6NCrl) have been sourced from Charles River Laboratories (UK). They have been housed beneath a typical 12 h mild, 12 h darkish cycle, with a temperature maintained between 20 and 24 °C, a relative humidity of 40–60% and have been monitored for indicators of regular well being, meals ingestion and body-weight achieve all through the research interval. In the beginning of the research, thirty-six mice (six weeks outdated) have been randomly divided into two feeding teams. As earlier than, one group acquired the usual rodent food regimen American Institute of Diet (AIN)−76A supplemented with 0.0625% (w/w) fgTiO2 (translating to roughly 100 mg fgTiO2 / kg body-weight/day respectively15), whereas the opposite acquired straight AIN-76A with out fgTiO2 supplementation (i.e., the unfavorable management food regimen). After sixteen weeks of feeding, all mice have been switched to the unfavorable management food regimen, and twelve mice from every diet-group have been orally challenged with ΔaroA-deficient, S. Typhimurium pressure SL326151 (ΔaroA–Salmonella). We marginally separated the administrations of micro organism and fgTiO2 to (i) forestall artefactual adsorption as a consequence of co-administration (ii) allow luminal interactions between gavaged micro organism and residual luminal particles (demonstrated by faecal ICP-MS research, Supplementary Fig. 16) ought to such interactions happen and (iii) be certain that some meals additionally was within the lumen beneath these circumstances as can be anticipated with regular feeding. Mice have been flippantly anaesthetised with isofluorane then orally gavaged with 0.2 mL of inoculum ready at 5.25 × 109 CFU/mL. Mice have been then euthanised by CO2 asphyxiation and cervical dislocation at two time-points, both +3 or +28 days after Salmonella inoculation (i.e., 16 weeks +3 days or 16 weeks +28 days). On this means, 4 therapy teams (n = 6 management food regimen, n = 12 management food regimen + Salmonella, n = 6 fgTiO2 food regimen, n = 12 fgTiO2 food regimen + Salmonella) have been established, with half of the animals analysed at every of the 2 time-points. Alongside, faecal samples have been collected at three time-points (pre-study, 16 weeks +3 days and 16 weeks +28 days) to allow enzyme-linked immunosorbent assay (ELISA) research for Salmonella-specific IgA and faecal Ti measurements by ICP-MS.
Human tissue assortment
Surgical specimens containing Peyer’s patches have been collected from the neo-terminal ileum from six sufferers with Crohn’s illness or colonic most cancers (3 males, 3 females; self-reported). These tissue samples have been made out there for microscopical evaluation with group-summary data solely (i.e., resection date-range 2017–2018; median age 48 years; age-range 21–77 years) so disaggregation of particular person microscopy knowledge by way of intercourse was not doable. In all cases the ileal tissue specimens shaped the resection margins and have been macroscopically regular. Peyer’s patches have been confirmed by means of microscopical identification52. The tissue specimens have been mounted with 4% PBS-buffered (pH 7.4) paraformaldehyde for 12 h at 4 °C earlier than immersion in 30% sucrose and freezing in OCT (LAMB/OCT, Thermo Scientific) in cryomolds. For microscopy analyses, the OCT-embedded tissue samples have been quickly shipped in a single day to Cambridge on dry ice. Alongside, three anonymised, formalin-fixed paraffin embedded (FFPE) human tissue samples containing ileal Peyer’s patch lymphoid tissue have been additionally analysed to verify if fgTiO2 may very well be detected within the FFPE specimen-type. Past tissue-type, no details about these human samples was out there.
Tissue sectioning and immunofluorescence labeling
Frozen sections have been minimize at 20-micron thickness utilizing a Leica CM 1900 cryostat, picked up on Superfrost Plus slides (ThermoFisher, J1800AMNT) and rested at room temperature for two h previous to labelling. FFPE sections have been minimize at 5-micron thickness, then dewaxed and rehydrated by baking at 60 °C for 1 h, transferring by means of two adjustments of xylene, a reverse ethanol collection (100%, 70%, 50%, 10%) adopted by 1 minute rehydration in water. Sections have been ringed with hydrophobic barrier pen (Vector Laboratories, H-4000) and cryostat sections mounted by publicity to recent 4%, 0.1 M PBS-buffered paraformaldehyde (pH 7.4) for 10 min. All subsequent steps have been performed beneath light agitation on a rotating shaker. To facilitate antibody penetration, sections have been permeabilised for 90 min utilizing 0.3 % (vol/vol) Triton X-100 in 0.1 M PBS (pH 7.4). Block buffer (10% goat serum, 2% bovine serum albumin, 20 mM glycine diluted in 25 mM Tris-buffered (pH 7.4) saline (TBS)) was then added to all sections for 1 h. Major antibodies or concentration-matched isotype controls have been diluted in block-buffer and incubated at 4 °C in a single day. All subsequent steps came about at room temperature. Sections have been washed with three adjustments of block-buffer. Secondary antibodies (the place directly-conjugated major antibodies weren’t used) have been diluted 1:400 in block buffer and incubated for 4 h. All antibody concentrations, fluorophore conjugations and producer data are offered in Supplementary Desk 2. 500 nM phalloidin-AlexaFluor 647 (ThermoFisher, A22287) and a couple of μg/mL Hoechst 33342 (ThermoFisher, H3570) have been additionally included alongside secondary antibodies to concurrently label cell outlines (i.e., cytoskeletal f-actin) and cell nuclei (respectively). Sections have been then washed as soon as with TBS previous to mounting with #1.5 coverslips in Delay Diamond (ThermoFisher, P36965).
Confocal microscopy & imaging controls
Fluorescence and reflectance image-data (2048×2048 pixels per tile) have been collected utilizing a diode-controlled (retrofitted) Zeiss LSM780 confocal microscope geared up with plan-apochromat 63X/1.4 and 40X/1.3 oil-immersion goals. Fluorescence and reflectance knowledge have been collected on the similar time throughout the identical imaging run by sequential scanning with mirrored (i.e., back-scattered) mild from a 488 nm excitation laser collected within the vary 485–491 nm to detect fgTiO215. The place tilescanning was used, photographs have been acquired with 10% edge overlap. Tiles have been stitched utilizing the Zen Black 2011 SP2 software program (Carl-Zeiss, UK) utilizing the nuclei and actin channels for registration and a strict correlation threshold setting of 1.0. For all datasets beneath comparability, photographs have been obtained in a single run utilizing similar settings. For the reflectance imaging, tissues from unfavorable management mice (i.e., fed the AIN-76 food regimen solely, so no fgTiO2 current) have been used to find out the picture acquisition settings and thresholding for fgTiO2 detection (technique proven, Supplementary Fig. 1). For immunofluorescence work, supporting secondary-only and concentration-matched isotype antibody controls in tissue-matched serial sections have been included. For datasets collected as units of single photographs from the villous mucosa (VM), sub-epithelial dome (SED), germinal heart (GC) and TCZ (T-cell zone) areas of mouse Peyer’s patches, landmarks (e.g., villi / follicle-associated epithelium) and T cell (CD3), B cell (B220) and mononuclear phagocyte (CD11c) immunofluorescence staining in adjoining sections have been used to tell imaging places. To outline the autofluorescence signature of the fgTiO2-recipient cells within the mouse Peyer’s patches, lambda scans have been carried out utilizing the LSM 780 microscope. Utilizing unstained tissue sections, autofluorescence upon excitation from a 405 nm laser was detected in ~10 nm intervals within the vary 415–700 nm (spectra offered, Supplementary Fig. 8).
Deconvolution
Z-stacked image-data (pixel measurement (XYZ) 1024x1024x123; voxel measurement (XYZ) 26x26x100 nm) of fgTiO2-recipient cells was collected utilizing a Zeiss LSM 780 confocal microscope geared up with a 63X/1.4 plan-apochromat oil immersion goal utilizing murine tissue sections embedded in Delay Glass (P36984, ThermoFisher) anti-fade reagent (refractive index ~ 1.5). Cell autofluorescence was excited utilizing the 405 nm laser. The image-stacks have been registered to right spatial drift utilizing StackReg53. The Born and Wolf scalar-based diffraction mannequin54 was then used to estimate a theoretical level unfold operate assuming a refractive index of 1.51 and a autofluorescence emission most of 495 nm (measured, Supplementary Fig. 8). The registered Z-stack was then deconvolved utilizing the Richardson-Lucy whole variation algorithm (75 iterations, regularisation 1e-12) throughout the open-source DeconvolutionLab255 software program.
fgTiO2 detection utilizing mirrored mild and circle-marker placement
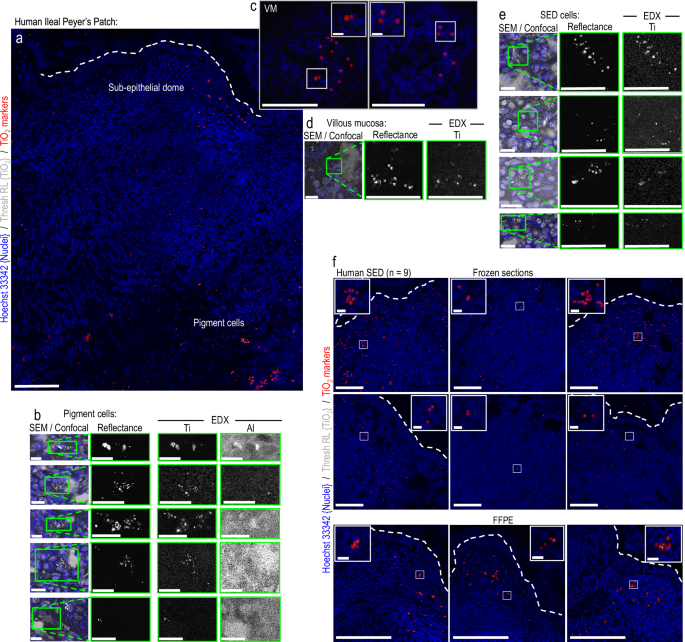
To allow fgTiO2 detection in organic tissue sections, mirrored mild and cell construction (i.e., nuclei / actin) fluorescence data have been collected on the similar time utilizing a Zeiss LSM780 confocal microscope. The reflectance photographs have been thresholded to isolate the punctate, reflectant foci brought on by the fgTiO2 particles from the background mild scatter brought on by the organic tissue. Because the background stage was comparable in each mouse and human specimens (proven, Supplementary Fig. 4) the identical threshold was used for each mouse and human photographs, and was decided as the extent required to take away all sign within the reflectance channel for photographs collected for tissue sections taken from the unfavorable management mice (i.e., fed the AIN-76 food regimen solely, so no fgTiO2 current). This strategy was then additional validated utilizing correlative scanning electron microscopy and power dispersive X-ray analyses to verify that the sign remaining after thresholding contained the anticipated X-ray sign for titanium (technique proven, Supplementary Fig. 1). Given the small measurement of the reflectant foci brought on by fgTiO2, a translucent circle-marker was positioned on every pixel above the brink utilizing the insertShape operate in MATLAB R2021a (MathWorks) to help visualisation given the comparatively small measurement of show within the Determine panels15. Examples of the uncooked sign and subsequent marker placements are proven for mouse and people in Supplementary Fig. 5. Instance knowledge and code demonstrating thresholding and circle marker placement can be found on the BioStudies database beneath accession quantity S-BSST875.
Correlative confocal-electron microscopy and power dispersive X-ray analyses
As soon as fgTiO2 detection by confocal reflectance microscopy was accomplished, Delay-mounted coverslips have been faraway from the tissue sections by in a single day incubation in PBS at 37 °C. Slides have been air dried at room temperature for twenty-four h, minimize down and mounted on aluminum SEM stubs utilizing colloidal graphite and coated with a skinny (~15 nm) layer of amorphous carbon (evaporative coater). Tissue sections have been then loaded right into a Helios G4 CX Twin Beam high-resolution, monochromated, discipline emission gun, scanning electron microscope (SEM) geared up with a exact centered ion beam (FIB) and in-lens secondary electron and round backscattered detectors (FEI/ThermoFisher). Secondary and backscattered electron photographs have been collected at 3–5 kV as tilescans for the whole tissue floor, allowing guide overlay onto the previously-collected confocal tilescans. To allow correlative confocal-SEM imaging, the define of the Peyer’s patch–significantly the follicle related epithelium and basal muscularis layer—have been manually registered collectively utilizing picture overlay, enabling triangulation to focus on particular areas containing reflectance-positive cells. The correlative confocal-to-EDX analyses of the human tissues have been then carried out instantly within the SEM, utilizing a 150 mm2 silicon drift detector and AZTEC software program (Oxford Devices). For the correlative scanning transmission electron microscopy (STEM) used to verify single-particle detection by reflectance microscopy, skinny lamella of the tissue pattern have been ready within the SEM by way of the in situ lift-out technique. As soon as sites-of-interest had been recognized, 500 nm of electron beam platinum (Pt) was deposited (at 5 kV, 6.4 nA for the electron supply) to the floor of the goal space. This was adopted by a second Pt layer (1 μm) utilizing the FIB (at 30 kV, 80 pA for the liquid gallium (Ga) ion supply). A bulk lamella was initially minimize utilizing the FIB (at 30 kV, 9 nA), earlier than the ultimate cut-out was carried out (at 30 kV, 80 pA). The lamella was connected, utilizing ion beam Pt, to a copper FIB lift-out grid (Omniprobe, USA) mounted throughout the SEM chamber (i.e., in situ). Last thinning and sharpening of the lamella to electron translucency was carried out with a mild polish/clear utilizing the ion beam (5 kV, 41 pA). A lamella was then transferred right into a Titan3 Themis G2 300 kV S/TEM geared up with an S-TWIN goal lens, monochromator, HAADF detector and a Tremendous-X 4-detector EDX system in a double-tilt specimen holder (FEI/ThermoFisher). Transmission electron microscopy (TEM) photographs and on-zone diffraction patterns have been recorded utilizing a OneView CMOS digicam (Gatan) and STEM-EDX elemental maps have been recorded and processed utilizing the Velox software program (FEI/ThermoFisher).
Cell segmentation of intestinal tissue photographs
In earlier work, we rigorously developed and validated staining and picture evaluation strategies enabling extraction of single-cell knowledge from confocal microscopy photographs of gastrointestinal tissues37. Based mostly on this framework, three cell segmentation pipelines have been used to accommodate completely different tissue varieties and differing availabilities of coaching knowledge. To substantiate the reliability of our previously-published pipelines with the image-data offered right here, the accuracy of the outputs from all three cell segmentation pipelines was assessed relative to manually-segmented cells by Jaccard index (offered, Supplementary Fig. 2).
Cell segmentation: mouse villous mucosa photographs
Villous image-fields have been instantly segmented into cell-objects utilizing the freely-available CellProfiler software program56. Utilizing the Hoechst 33342 fluorescence data, nuclei have been first segmented as major objects. These have been then used as seeds throughout a marker-controlled watershed course of to phase every cell’s define utilizing fluorescence data from the actin channel. The method is schematically demonstrated in Supplementary Fig. 17 with the whole CellProfiler pipeline proven in Supplementary Notice 1. Take a look at photographs and the CellProfiler pipeline are additionally out there for obtain on the BioStudies database beneath accession quantity S-BSST875.
Cell segmentation: mouse lymphoid tissue photographs
Lymphoid tissues (e.g., Peyer’s patches and caecal patches) exhibit excessive cell densities resulting in poor cell segmentation accuracies utilizing the easy, marker-controlled watershed strategy used for the villous mucosa photographs37. Pixel-classification machine studying can be utilized to alleviate this drawback by means of the era of chance photographs that extra exactly delineate every cell’s boundary37. Right here, in mouse lymphoid tissues, these chance photographs have been offered utilizing a 2-D U-Internet absolutely convolutional neural community57 deployed in MATLAB R2021a (MathWorks, MA). The community has a 256x256x2 (x, y, channels) enter layer and makes use of the nuclear and actin fluorescence data to foretell the chance that every pixel belongs to both cell define, intracellular atmosphere or background classifications. The coaching knowledge consisted of twelve lymphoid tissues photographs (every containing 2138 × 1900 annotated pixels overlaying ~16,000 cells ready by an skilled cell biologist). The image-data flows by means of a four-layer contracting convolutional path earlier than full up-convolutional enlargement, yielding chance photographs that precisely match the input-image dimensions. The community was skilled for 50 epochs utilizing a batch-size of 12 for 1200 iterations/epoch. Patches have been shuffled each epoch and augmented by easy, random x/y reflection and rotation. Mannequin coaching was optimised beneath stochastic gradient descent utilizing cross-entropy loss. The preliminary studying price was 0.05, dropping each 10 epochs by 0.1 beneath momentum 0.9 and L2 regularisation 1 × 10−4. The resultant cell define chance photographs have been then loaded into the CellProfiler pipeline alongside immunofluorescence and reflectance data collected by the confocal microscope and have been used to allow segmentation of particular person cells by way of an IdentifyPrimaryObjects module. This course of is schematically demonstrated in Supplementary Fig. 18. The entire CellProfiler picture evaluation pipeline is offered in Supplementary Notice 2. All U-Internet code, the skilled community with test-data and the whole CellProfiler pipeline can be found for obtain from the BioStudies database beneath accession quantity S-BSST875.
Cell/TLV segmentation: correlative mouse-human dosimetry
For the correlative analyses investigating the relative quantities of fgTiO2 delivered to particular person SED cells in mice versus in normally-exposed people, frozen ileal tissue sections containing Peyer’s patches have been collected at random from six normally-exposed people, and 6 mice fed the AIN-67 food regimen supplemented with 0.0625% (w/w) fgTiO2. In each occasion, care was taken to gather tissue sections with clear follicle-associated epithelium (i.e., with none overlying villi) to make sure upkeep of comparable histological positioning close to the middle of the dome of every Peyer’s patch. Reflectance confocal photographs of the murine and human tissues have been then acquired in a single run beneath similar settings. After acquisition, the image-fields per specimen have been manually specified by order of lowest-to-highest fgTiO2 accumulation to visually current the spectrum of (extensive) variation in fgTiO2 mobile loading noticed throughout samples in each the mouse and human tissue specimens. This was adopted by quantitative picture evaluation to find out the thresholded reflectance per unit tissue space; then the built-in depth of the brink reflectance data per-cell and per-vesicle objects (technique exemplified, Supplementary Fig. 3). To extract the single-cell and single-vesicle knowledge, a CellProfiler pipeline was used. To deal with cell segmentation in lymphoid tissues with excessive cell densities we’ve proven in earlier work37 that pixel-classification machine studying can be utilized to supply chance photographs that extra exactly delineate every cell’s boundary enhancing segmentation accuracies when in comparison with utilizing the uncooked fluorescence data. In contrast to in mouse lymphoid tissues (technique described above), as a result of giant quantities of manually-annotated coaching knowledge for the human lymphoid photographs weren’t out there, these chance photographs have been created by sparse annotation pixel-classification machine studying within the ILASTIK software program58. Two ILASTIK pixel-classification tasks have been created (one for the human photographs, one for the mouse). Pixel annotations representing intracellular atmosphere, cell outlines and different classifications have been made on the fluorescence channel describing the cell outlines (i.e., the actin channel) (technique proven schematically, Supplementary Fig. 19). The ILASTIK software program then generated chance photographs representing the chance of every pixel belonging to every classification. The resultant cell define chance photographs have been loaded into the CellProfiler pipeline alongside the fluorescence and reflectance data captured by the confocal microscope and have been used to phase particular person cells by way of an IdentifyPrimaryObjects module. As a part of this pipeline, the reflectant foci representing fgTiO2 occasions have been additionally directly-segmented from the thresholded reflectance photographs into fgTiO2-loaded vesicle (TLV) objects utilizing a second, IdentifyPrimaryObjects module (technique schematically demonstrated, Supplementary Fig. 20). The entire picture evaluation pipeline is offered in Supplementary Notice 3. Take a look at photographs and the whole CellProfiler pipeline can be found for obtain from the BioStudies database beneath accession quantity S-BSST875. The accuracy of the segmented-cell outputs was checked by Jaccard index relative to manually-segmented cells (offered, Supplementary Fig. 2). The accuracy of TLV segmentation from the uncooked reflectance data is explored in each mouse and human photographs in Supplementary Fig. 5.
Cell/TLV function extraction
Uncooked immunofluorescence knowledge have been pre-processed by guide thresholding on the stage required to take away ≥95% of fluorescence in tissue-matched, secondary antibody-only management photographs37. Alongside, the reflectance knowledge have been rigorously thresholded on the stage required to take away all sign from the pictures collected from the unfavorable management group mice (e.g., fed the AIN-67 food regimen alone; technique proven, Supplementary Fig. 1). Thresholded reflectance and fluorescence depth data per cell or TLV object, alongside measurement and form options have been then measured for each the immunofluorescence and reflectance channels utilizing the MeasureObjectSizeShape and MeasureObjectIntensity modules in CellProfiler. The variety of TLV objects per cell was measured utilizing a RelateObjects module. Knowledge have been preprocessed by discarding objects outdoors of the fifth and ninety fifth percentiles by measurement previous to evaluation as is really useful best-practice59.
Cell segmentation: block processing tilescans
To keep away from reminiscence limitations, when vital, tilescanned photographs have been minimize into smaller items, run by means of one of many CellProfiler pipelines earlier than reassembling the item segmentation masks and assigning world cell place coordinates to the extracted object options. This was achieved utilizing two MATLAB features developed in our earlier work37. The primary TilescanToCellProfiler, reads stitched photographs in most microscopy codecs and cuts them into manageable tiles with edge-overlap for processing. The second, CellProfilerToTilescan reassembles the item masks, removes double hits on overlap edges and assigns world place coordinates values to the extracted options. Instance photographs and MATLAB code demonstrating these features can be found for obtain on the BioStudies database beneath accession quantity S-BSST875.
Cell segmentation: accuracy evaluation by Jaccard index
The Jaccard index (intersection over union) strategy was used to test the accuracy of the cell segmentation outcomes used on this work. This was achieved by evaluating pixel positions contained in the automatically-segmented cell objects in opposition to these obtained by guide annotations carried out by an expertise cell biologist. The Jaccard index was calculated as:
$$Jleft(A,Mright)=frac{Acap }{Acup }=frac{Acap }-Acap $$
(1)
The place J is the Jaccard distance for 2 units of pixel positions for the automated (A) and guide (M) segmentations, respectively. Scores of zero signify no overlap (false negatives) whereas scores of 1 signify actual pixel-for-pixel intersection. On this means, it’s acknowledged that this strategy is a comparatively harsh success measure, and that scores of ~0.7 point out a superb segmentation consequence57. That is partially because of the inevitable inaccuracies which can be current—even within the manually-annotated knowledge (e.g., as a consequence of define smoothing, ambiguity in figuring out the exact place of every cell’s boundary from the fluorescent staining data, and the out there picture decision, and so forth.).
Cell segmentation: cell mapping visualisations
The cell map visualisations displaying the delivered mobile doses of fgTiO2 or TLV counts per-cell have been produced utilizing cell function data extracted by the CellProfiler MeasureObjectIntensities and RelateObjects modules alongside the item segmentation masks outputted by every pipeline. Utilizing MATLAB scripts, object function knowledge have been binned into classes (e.g., low, intermediate and excessive fgTiO2-load primarily based on the built-in depth of the thresholded reflectance sign per-cell). Utilizing the segmentation masks, particular person cell-objects have been then colour-coded in response to these classes to offer a tissue map visualisation. Instance knowledge and MATLAB code demonstrating the creation of those visualisations can be found for obtain from the BioStudies database beneath accession quantity S-BSST875.
Statistical evaluation of image-based knowledge
Built-in depth per object (i.e., per cell or NLV) distributions have been in contrast utilizing two-sided Wilcoxon rank-sum evaluation. This non-parametric strategy was chosen for compatibility with unpaired samples with unequal group sizes37. The strategy exams the null speculation that the distributions beneath comparability would possibly moderately be drawn from one steady distribution with equal medians (P > 0.05) versus the choice speculation that the distributions are distinct (P ≤ 0.05). Pairwise comparisons of cell counts have been first checked for distribution normality and variance homogeneity utilizing Shapiro-Wilk and Bartlett exams, respectively. The place these exams have been handed (P > 0.05), teams have been in contrast utilizing a two-sided, unpaired samples T-test assuming equal variance.
Olink® proximity extension assay
Frozen mouse ileal samples have been weighed and saved in Kimble pellet pestle tubes (DWK Life Sciences, Okay-749520-0000) at −70 °C till processing. The Peyer’s patch-enriched samples have been first trimmed tightly to both facet of Peyer’s patch utilizing a chilly security razorblade such that every pattern primarily consisted of solely the patch itself. Tissues have been lysed utilizing the BioPlex Cell Lysis Package (BioRad, 171-304011). 500 μL of Cell Wash Buffer per pattern was used for rinsing the tissues. Cell Lysis Buffer was ready by including Components 1 and a couple of as per the producer’s directions, in addition to Halt Protease Inhibitor Cocktail (ThermoFisher, 78437; 10 μL of cocktail per 1 mL of buffer). Following a rinse, tissues have been homogenised by including 500 μL of ice-cold Cell Lysis Buffer and hand grinding on ice utilizing disposable pestles (DWK Life Sciences, 749520-0000; 20 strokes). Homogenised samples have been frozen at −70 °C for at the least 4 h, thawed on ice, sonicated in an Ultrawave U300 ultrasonic bathtub (44 kHz, 35 W) on ice for 40 seconds and centrifuged for five min at 6000 × g. Supernatant was collected into 2 mL cryogenic vials (Corning, 430659) and frozen at −70 °C till quantification. Quantification was carried out utilizing the DC Protein Quantification Package I (BioRad, 5000111) as per the producer’s directions (Microplate Assay Protocol). Plates (Greiner Bio-One, 650185) have been learn utilizing FLUOstar Omega microplate reader at 750 nm and protein concentrations have been calculated by the Omega’s MARS Knowledge Evaluation software program utilizing linear regression match. All samples have been normalised to 0.77 mg/mL (+/− 0.12) by dilution with Cell Lysis Buffer and re-quantified to verify protein focus. A microplate was ready for the proximity extension assay by transferring 40 μL aliquots of all samples, unfavorable controls (Cell Lysis Buffer), duplicates of 6 samples, and dilutions (1:4, 1:8 and 1:16 v/v) of two samples right into a 96-well skirted plate (ThermoFisher, AB0800) in a randomised method (controls offered, Supplementary Fig. 11). The plate was sealed (ThermoFisher, 4306311) and saved at −70 °C till cargo on dry ice to Olink (Uppsala, Sweden) for evaluation utilizing the Goal 96 Mouse Exploratory panel (www.olink.com/mouse-exploratory). All protein names and abbreviations are listed in Supplementary Desk 1. Statistical evaluation was carried out utilizing the Olink Analyze 3.6.0 R-package (https://CRAN.R-project.org/package deal=OlinkAnalyze). Therapy teams have been in contrast utilizing a two-sample Mann-Whitney U take a look at with correction for a number of testing utilizing the Benjamini-Hochberg technique. In the principle textual content, outcomes for proteins that may very well be concerned in inflammatory signaling are proven (Fig. 5) whereas within the Supplementary Info outcomes of your entire protein set are proven alongside repeat-sample, serial dilution and clean pattern proximity extension assay controls (Supplementary Figs. 11–13). The proximity extension assay knowledge can be found for obtain on the BioStudies database beneath accession quantity S-BSST875.
Enzyme-linked immunosorbent assay for Salmonella-specific IgA
Throughout Mouse Research 2, faecal pellets have been collected pre-study and on the 16-week +3 day and 16-week +28 day time-points. Samples have been weighed and diluted 1:10 (w/v) in 10% PBS containing 1 mM EDTA. The ensuing suspension was vortexed for 15 min to facilitate liquefaction then centrifuged at 400 x g at 4 °C for five min. The supernatant was aspirated and centrifuged for a second time at 12,000 x g for 10 min at 4 °C. Salmonella-specific IgA quantification was then carried by sandwich ELISA technique. Plates have been coated with 50 μL lipopolysaccharide (LPS) (Sigma, L2262) diluted in Reggiardo’s buffer (0.05 M glycine, 0.1 M NaCl, 1 mM EDTA, 0.05 M NaF and 0.1% sodium deoxycholate). Plates have been then sealed and incubated in a single day at 37 °C. Subsequent day, the plates have been washed 3 instances with PBS-tween and blotted dry. Blocking buffer from the entire IgA equipment (Affymetrix, 88-50450) was added to every effectively for 1 h at 37 °C. The faecal supernatant was then diluted in equipment buffer A (Affymetrix, 88-50450) and 50 μL of diluted pattern was added to every effectively. After 3 h of incubation, the plates have been washed thrice with PBS-tween earlier than addition of fifty μL horseradish peroxidase-conjugated IgA. Three extra washes have been carried out earlier than the addition of fifty μL tetramethylbenzidine substrate for 10 min. This was adopted by 50 μL of cease resolution previous to absorption studying at 492 nm on a plate reader. All outputs have been calibrated utilizing a typical curve and corrected for dilution.
Faecal ICP-MS evaluation
Faecal samples have been collected and saved at −80 °C. Samples have been weighed and a digestion resolution of 1:1 (vol/vol) nitric acid (Fisher, 7697-37-2) and hydrogen peroxide (Sigma, 7722-84-1) was added to the samples (20–70 mg faecal weight) at a 1:10 (wt/vol) ratio. Samples have been digested at room temperature for 72 h earlier than loosely-capped pattern containers have been transferred to a water bathtub (40 °C) for five–6 h to launch the peroxide. Samples have been then transferred to a PTFE vial and sulphuric acid was added (1:1 vol acid/pattern weight) previous to closing digestion utilizing a Milestone UltraWave microwave. The resultant liquid was decanted and diluted 1:10 (vol/vol) utilizing ultra-high purity water. Quantification of elemental Ti was carried out utilizing an 8900 triple quadruple inductively coupled plasma mass spectrometer (Agilent, USA). Scans have been carried out in MS/MS mode with the response cell adjusted for evaluation in H2/O2 fuel. The instrument was set as much as establish the mass pairs 48 and 64 for isotopic Ti-48 and TiO2-64. Calibration requirements have been ready in 2% nitric acid and spiked with elemental Ti (Sigma, 12237) to closing concentrations between 0 and 1 ppm. All samples have been spiked with an inner normal resolution containing gallium (Sigma, 16639), germanium (Sigma, 05419), yttrium (Sigma, 01357) & europium (Sigma, 05779) at closing concentrations of two ppb. Counts per-second (cps) values for yttrium have been collected to reference sign stability. A 2% resolution of nitric acid was used as pattern provider and rinse.
Reporting abstract
Additional data on analysis design is accessible within the Nature Portfolio Reporting Abstract linked to this text.